Heterochromatin-Encoded Satellite RNAs Induce Breast Cancer
- PMID: 29861157
- PMCID: PMC6545586
- DOI: 10.1016/j.molcel.2018.04.023
Heterochromatin-Encoded Satellite RNAs Induce Breast Cancer
Abstract
Heterochromatic repetitive satellite RNAs are extensively transcribed in a variety of human cancers, including BRCA1 mutant breast cancer. Aberrant expression of satellite RNAs in cultured cells induces the DNA damage response, activates cell cycle checkpoints, and causes defects in chromosome segregation. However, the mechanism by which satellite RNA expression leads to genomic instability is not well understood. Here we provide evidence that increased levels of satellite RNAs in mammary glands induce tumor formation in mice. Using mass spectrometry, we further show that genomic instability induced by satellite RNAs occurs through interactions with BRCA1-associated protein networks required for the stabilization of DNA replication forks. Additionally, de-stabilized replication forks likely promote the formation of RNA-DNA hybrids in cells expressing satellite RNAs. These studies lay the foundation for developing novel therapeutic strategies that block the effects of non-coding satellite RNAs in cancer cells.
Keywords: BRCA1-associated proteins; DNA damage response; breast tumor formation; heterochromatic RNAs; satellite RNAs.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures

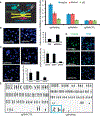



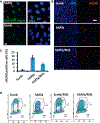
References
-
- Aguilera A, and Gómez-González B (2008). Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 9, 204–217. - PubMed
-
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. (2004). Global identification of human transcribed sequences with genome tiling arrays. Science 306,2242–2246. - PubMed
-
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al.; ENCODE Project Consortium; NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center; Washington University Genome Sequencing Center; Broad Institute; Children’s Hospital Oakland Research Institute (2007). Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

