Large set data mining reveals overexpressed GPCRs in prostate and breast cancer: potential for active targeting with engineered anti-cancer nanomedicines
- PMID: 29861840
- PMCID: PMC5982759
- DOI: 10.18632/oncotarget.25427
Large set data mining reveals overexpressed GPCRs in prostate and breast cancer: potential for active targeting with engineered anti-cancer nanomedicines
Abstract
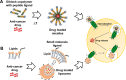
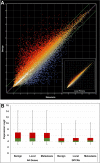
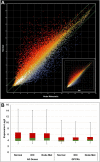
Over 800 G-protein-coupled receptors (GPCRs) are encoded by the human genome and many are overexpressed in tumors. GPCRs are triggered by ligand molecules outside the cell and activate internal signal transduction pathways driving cellular responses. The receptor signals are desensitized by receptor internalization and this mechanism can be exploited for the specific delivery of ligand-linked drug molecules directly into cells. Detailed expression analysis in cancer tissue can inform the design of GPCR-ligand decorated drug carriers for active tumor cell targeting. The active targeting process utilizes ligand receptor interactions leading to binding and in most cases internalization of the ligand-attached drug carrier resulting in effective targeting of cancer cells. In this report public microarray data from the Gene Expression Omnibus (GEO) repository was used to identify overexpressed GPCRs in prostate and breast cancer tissues. The analyzed data confirmed previously known cancer receptor associations and identified novel candidates for potential active targeting. Prioritization of the identified targeting receptors is also presented based on high expression levels and frequencies in cancer samples but low expression in healthy tissue. Finally, some selected examples were used in ligand docking studies to assess the feasibility for chemical conjugation to drug nanocarriers without interference of receptor binding and activation. The presented data demonstrate a large untapped potential to improve efficacy and safety of current and future anti-cancer compounds through active targeting of GPCRs on cancer cells.
Keywords: GPCR; Pathology; cancer; meta-data; nanocarrier; targeted chemotherapy.
Conflict of interest statement
CONFLICTS OF INTEREST The authors report no conflicts of interest in this work.
Figures
References
-
- Wicki A, Witzigmann D, Balasubramanian V, Huwyler J. Nanomedicine in cancer therapy: challenges, opportunities, and clinical applications. J Control Release. 2015;200:138–57. - PubMed
-
- Gaumet M, Vargas A, Gurny R, Delie F. Nanoparticles for drug delivery: the need for precision in reporting particle size parameters. Eur J Pharm Biopharm. 2008;69:1–9. - PubMed
-
- Barragan F, Carrion-Salip D, Gomez-Pinto I, Gonzalez-Canto A, Sadler PJ, de Llorens R, Moreno V, Gonzalez C, Massaguer A, Marchan V. Somatostatin subtype-2 receptor-targeted metal-based anticancer complexes. Bioconjug Chem. 2012;23:1838–55. - PubMed
-
- de Jong M, Breeman WA, Kwekkeboom DJ, Valkema R, Krenning EP. Tumor imaging and therapy using radiolabeled somatostatin analogues. Acc Chem Res. 2009;42:873–80. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

