Clinotator: analyzing ClinVar variation reports to prioritize reclassification efforts
- PMID: 29862020
- PMCID: PMC5941247
- DOI: 10.12688/f1000research.14470.2
Clinotator: analyzing ClinVar variation reports to prioritize reclassification efforts
Abstract
While ClinVar has become an indispensable resource for clinical variant interpretation, its sophisticated structure provides it with a daunting learning curve. Often the sheer depth of types of information provided can make it difficult to analyze variant information with high throughput. Clinotator is a fast and lightweight tool to extract important aspects of criteria-based clinical assertions; it uses that information to generate several metrics to assess the strength and consistency of the evidence supporting the variant clinical significance. Clinical assertions are weighted by significance type, age of submission and submitter expertise category to filter outdated or incomplete assertions that otherwise confound interpretation. This can be accomplished in batches: either lists of Variation IDs or dbSNP rsIDs, or with vcf files that are additionally annotated. Using sample sets ranging from 15,000-50,000 variants, we slice out problem variants in minutes without extensive computational effort (using only a personal computer) and corroborate recently reported trends of discordance hiding amongst the curated masses. With the rapidly growing body of variant evidence, most submitters and researchers have limited resources to devote to variant curation. Clinotator provides efficient, systematic prioritization of discordant variants in need of reclassification. The hope is that this tool can inform ClinVar curation and encourage submitters to keep their clinical assertions current by focusing their efforts. Additionally, researchers can utilize new metrics to analyze variants of interest in pursuit of new insights into pathogenicity.
Keywords: ClinVar; benign; clinical variant; pathogenic; pathogenicity; variant interpretation; variant reclassification; variation.
Conflict of interest statement
No competing interests were disclosed.
Figures
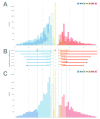
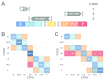
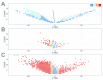

References
-
- Richards S, Aziz N, Bale S, et al. : Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. 10.1038/gim.2015.30 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

