Long read assemblies of geographically dispersed Plasmodium falciparum isolates reveal highly structured subtelomeres
- PMID: 29862326
- PMCID: PMC5964635
- DOI: 10.12688/wellcomeopenres.14571.1
Long read assemblies of geographically dispersed Plasmodium falciparum isolates reveal highly structured subtelomeres
Abstract
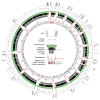
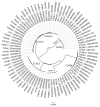
Background: Although thousands of clinical isolates of Plasmodium falciparum are being sequenced and analysed by short read technology, the data do not resolve the highly variable subtelomeric regions of the genomes that contain polymorphic gene families involved in immune evasion and pathogenesis. There is also no current standard definition of the boundaries of these variable subtelomeric regions. Methods: Using long-read sequence data (Pacific Biosciences SMRT technology), we assembled and annotated the genomes of 15 P. falciparum isolates, ten of which are newly cultured clinical isolates. We performed comparative analysis of the entire genome with particular emphasis on the subtelomeric regions and the internal var genes clusters. Results: The nearly complete sequence of these 15 isolates has enabled us to define a highly conserved core genome, to delineate the boundaries of the subtelomeric regions, and to compare these across isolates. We found highly structured variable regions in the genome. Some exported gene families purportedly involved in release of merozoites show copy number variation. As an example of ongoing genome evolution, we found a novel CLAG gene in six isolates. We also found a novel gene that was relatively enriched in the South East Asian isolates compared to those from Africa. Conclusions: These 15 manually curated new reference genome sequences with their nearly complete subtelomeric regions and fully assembled genes are an important new resource for the malaria research community. We report the overall conserved structure and pattern of important gene families and the more clearly defined subtelomeric regions.
Keywords: Long read Assembly; Plasmodium falciparum; complete genomes; definition of core genome.
Conflict of interest statement
No competing interests were disclosed.
Figures








References
-
- WHO: Fact Sheet: World Malaria Day 2016.2016. Reference Source
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

