Gastrointestinal stromal tumor enhancers support a transcription factor network predictive of clinical outcome
- PMID: 29866822
- PMCID: PMC6016782
- DOI: 10.1073/pnas.1802079115
Gastrointestinal stromal tumor enhancers support a transcription factor network predictive of clinical outcome
Abstract
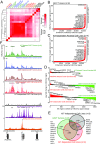
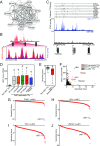
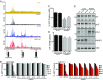
Activating mutations in the KIT or PDGFRA receptor tyrosine kinases are hallmarks of gastrointestinal stromal tumor (GIST). The biological underpinnings of recurrence following resection or disease progression beyond kinase mutation are poorly understood. Utilizing chromatin immunoprecipitation with sequencing of tumor samples and cell lines, we describe the enhancer landscape of GIST, highlighting genes that reinforce and extend our understanding of these neoplasms. A group of core transcription factors can be distinguished from others unique to localized and metastatic disease. The transcription factor HAND1 emerges in metastatic disease, binds to established GIST-associated enhancers, and facilitates GIST cell proliferation and KIT gene expression. The pattern of transcription factor expression in primary tumors is predictive of metastasis-free survival in GIST patients. These results provide insight into the enhancer landscape and transcription factor network underlying GIST, and define a unique strategy for predicting clinical behavior of this disease.
Keywords: GIST; epigenetics; sarcoma; transcription factor.
Conflict of interest statement
Conflict of interest statement: R.Z. is currently an employee at C4 Therapeutics. S.A.A. is a consultant for Epizyme Inc., Vitae Inc., and Imago Biosciences. G.D.D. reports financial relationships with Ariad, Astra-Zeneca, Bayer, Blueprint Medicines, Kolltan Pharmaceuticals, and Pfizer. J.E.B. is an employee, shareholder, and executive of Novartis Pharmaceuticals. J.E.B. is a scientific founder of Syros Pharmaceuticals, SHAPE Pharmaceuticals, Acetylon Pharmaceuticals, Tensha Therapeutics (now Roche), and C4 Therapeutics, and is the inventor on intellectual property licensed to these entities. None of these relationships constitute a conflict of interest for the present work. The remaining authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

