A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure
- PMID: 29866852
- PMCID: PMC6016793
- DOI: 10.1073/pnas.1802429115
A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure
Abstract
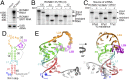
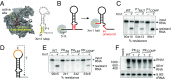
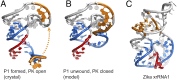
Folded RNA elements that block processive 5' → 3' cellular exoribonucleases (xrRNAs) to produce biologically active viral noncoding RNAs have been discovered in flaviviruses, potentially revealing a new mode of RNA maturation. However, whether this RNA structure-dependent mechanism exists elsewhere and, if so, whether a singular RNA fold is required, have been unclear. Here we demonstrate the existence of authentic RNA structure-dependent xrRNAs in dianthoviruses, plant-infecting viruses unrelated to animal-infecting flaviviruses. These xrRNAs have no sequence similarity to known xrRNAs; thus, we used a combination of biochemistry and virology to characterize their sequence requirements and mechanism of stopping exoribonucleases. By solving the structure of a dianthovirus xrRNA by X-ray crystallography, we reveal a complex fold that is very different from that of the flavivirus xrRNAs. However, both versions of xrRNAs contain a unique topological feature, a pseudoknot that creates a protective ring around the 5' end of the RNA structure; this may be a defining structural feature of xrRNAs. Single-molecule FRET experiments reveal that the dianthovirus xrRNAs undergo conformational changes and can use "codegradational remodeling," exploiting the exoribonucleases' degradation-linked helicase activity to help form their resistant structure; such a mechanism has not previously been reported. Convergent evolution has created RNA structure-dependent exoribonuclease resistance in different contexts, which establishes it as a general RNA maturation mechanism and defines xrRNAs as an authentic functional class of RNAs.
Keywords: RNA dynamics; RNA structure; exoribonuclease resistance; noncoding RNA maturation; single-molecule FRET.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Jones CI, Zabolotskaya MV, Newbury SF. The 5′ → 3′ exoribonuclease XRN1/Pacman and its functions in cellular processes and development. Wiley Interdiscip Rev RNA. 2012;3:455–468. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

