Altered p53 functionality in cancer-associated fibroblasts contributes to their cancer-supporting features
- PMID: 29866855
- PMCID: PMC6016816
- DOI: 10.1073/pnas.1719076115
Altered p53 functionality in cancer-associated fibroblasts contributes to their cancer-supporting features
Abstract
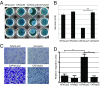
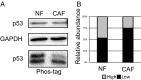
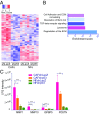
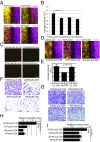
Within the tumor microenvironment, cancer cells coexist with noncancerous adjacent cells that constitute the tumor microenvironment and impact tumor growth through diverse mechanisms. In particular, cancer-associated fibroblasts (CAFs) promote tumor progression in multiple ways. Earlier studies have revealed that in normal fibroblasts (NFs), p53 plays a cell nonautonomous tumor-suppressive role to restrict tumor growth. We now wished to investigate the role of p53 in CAFs. Remarkably, we found that the transcriptional program supported by p53 is altered substantially in CAFs relative to NFs. In agreement, the p53-dependent secretome is also altered in CAFs. This transcriptional rewiring renders p53 a significant contributor to the distinct intrinsic features of CAFs, as well as promotes tumor cell migration and invasion in culture. Concordantly, the ability of CAFs to promote tumor growth in mice is greatly compromised by depletion of their endogenous p53. Furthermore, cocultivation of NFs with cancer cells renders their p53-dependent transcriptome partially more similar to that of CAFs. Our findings raise the intriguing possibility that tumor progression may entail a nonmutational conversion ("education") of stromal p53, from tumor suppressive to tumor supportive.
Keywords: fibroblasts; lung cancer; matrix metalloproteinases; p53; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

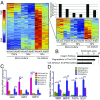
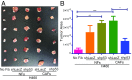
References
-
- Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

