Spatial transcriptomic survey of human embryonic cerebral cortex by single-cell RNA-seq analysis
- PMID: 29867213
- PMCID: PMC6028726
- DOI: 10.1038/s41422-018-0053-3
Spatial transcriptomic survey of human embryonic cerebral cortex by single-cell RNA-seq analysis
Abstract
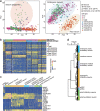
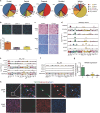
The cellular complexity of human brain development has been intensively investigated, although a regional characterization of the entire human cerebral cortex based on single-cell transcriptome analysis has not been reported. Here, we performed RNA-seq on over 4,000 individual cells from 22 brain regions of human mid-gestation embryos. We identified 29 cell sub-clusters, which showed different proportions in each region and the pons showed especially high percentage of astrocytes. Embryonic neurons were not as diverse as adult neurons, although they possessed important features of their destinies in adults. Neuron development was unsynchronized in the cerebral cortex, as dorsal regions appeared to be more mature than ventral regions at this stage. Region-specific genes were comprehensively identified in each neuronal sub-cluster, and a large proportion of these genes were neural disease related. Our results present a systematic landscape of the regionalized gene expression and neuron maturation of the human cerebral cortex.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

