The Immediate Early Gene Egr3 Is Required for Hippocampal Induction of Bdnf by Electroconvulsive Stimulation
- PMID: 29867393
- PMCID: PMC5958205
- DOI: 10.3389/fnbeh.2018.00092
The Immediate Early Gene Egr3 Is Required for Hippocampal Induction of Bdnf by Electroconvulsive Stimulation
Abstract
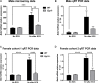
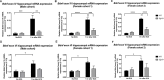
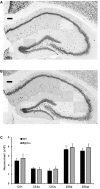
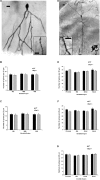
Early growth response 3 (Egr3) is an immediate early gene (IEG) that is regulated downstream of a cascade of genes associated with risk for psychiatric disorders, and dysfunction of Egr3 itself has been implicated in schizophrenia, bipolar disorder, and depression. As an activity-dependent transcription factor, EGR3 is poised to regulate the neuronal expression of target genes in response to environmental events. In the current study, we sought to identify a downstream target of EGR3 with the goal of further elucidating genes in this biological pathway relevant for psychiatric illness risk. We used electroconvulsive stimulation (ECS) to induce high-level expression of IEGs in the brain, and conducted expression microarray to identify genes differentially regulated in the hippocampus of Egr3-deficient (-/-) mice compared to their wildtype (WT) littermates. Our results replicated previous work showing that ECS induces high-level expression of the brain-derived neurotrophic factor (Bdnf) in the hippocampus of WT mice. However, we found that this induction is absent in Egr3-/- mice. Quantitative real-time PCR (qRT-PCR) validated the microarray results (performed in males) and replicated the findings in two separate cohorts of female mice. Follow-up studies of activity-dependent Bdnf exons demonstrated that ECS-induced expression of both exons IV and VI requires Egr3. In situ hybridization demonstrated high-level cellular expression of Bdnf in the hippocampal dentate gyrus following ECS in WT, but not Egr3-/-, mice. Bdnf promoter analysis revealed eight putative EGR3 binding sites in the Bdnf promoter, suggesting a mechanism through which EGR3 may directly regulate Bdnf gene expression. These findings do not appear to result from a defect in the development of hippocampal neurons in Egr3-/- mice, as cell counts in tissue sections stained with anti-NeuN antibodies, a neuron-specific marker, did not differ between Egr3-/- and WT mice. In addition, Sholl analysis and counts of dendritic spines in golgi-stained hippocampal sections revealed no difference in dendritic morphology or synaptic spine density in Egr3-/-, compared to WT, mice. These findings indicate that Egr3 is required for ECS-induced expression of Bdnf in the hippocampus and suggest that Bdnf may be a downstream gene in our previously identified biologically pathway for psychiatric illness susceptibility.
Keywords: brain-derived neurotrophic factor; early growth response 3; electroconvulsive therapy; immediate early genes; psychosis treatment; schizophrenia.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

