A Computational Workflow for Probabilistic Quantitative in Vitro to in Vivo Extrapolation
- PMID: 29867507
- PMCID: PMC5968095
- DOI: 10.3389/fphar.2018.00508
A Computational Workflow for Probabilistic Quantitative in Vitro to in Vivo Extrapolation
Abstract
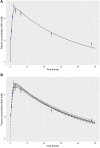
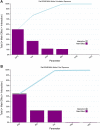
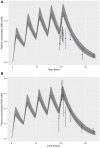
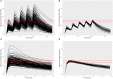
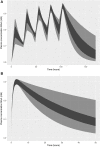
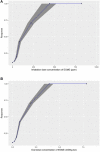
A computational workflow was developed to facilitate the process of quantitative in vitro to in vivo extrapolation (QIVIVE), specifically the translation of in vitro concentration-response to in vivo dose-response relationships and subsequent derivation of a benchmark dose value (BMD). The workflow integrates physiologically based pharmacokinetic (PBPK) modeling; global sensitivity analysis (GSA), Approximate Bayesian Computation (ABC) and Markov Chain Monte Carlo (MCMC) simulation. For a given set of in vitro concentration and response data the algorithm returns the posterior distribution of the corresponding in vivo, population-based dose-response values, for a given route of exposure. The novel aspect of the workflow is a rigorous statistical framework for accommodating uncertainty in both the parameters of the PBPK model (both parameter uncertainty and population variability) and in the structure of the PBPK model itself recognizing that the model is an approximation to reality. Both these sources of uncertainty propagate through the workflow and are quantified within the posterior distribution of in vivo dose for a fixed representative in vitro concentration. To demonstrate this process and for comparative purposes a similar exercise to previously published work describing the kinetics of ethylene glycol monoethyl ether (EGME) and its embryotoxic metabolite methoxyacetic acid (MAA) in rats was undertaken. The computational algorithm can be used to extrapolate from in vitro data to any organism, including human. Ultimately, this process will be incorporated into a user-friendly, freely available modeling platform, currently under development, that will simplify the process of QIVIVE.
Keywords: PBPK; benchmark dose; computational; extrapolation; in vitro; in vivo; workflow.
Figures

References
-
- Bahinski A. (2015). The Promise and Potential of “Organs-on-Chips” as Preclinical Models. Applied In Vitro Toxicology 1, 235–242. 10.1089/aivt.2015.29002.rtl - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

