Coptisine Induces Apoptosis in Human Hepatoma Cells Through Activating 67-kDa Laminin Receptor/cGMP Signaling
- PMID: 29867512
- PMCID: PMC5968218
- DOI: 10.3389/fphar.2018.00517
Coptisine Induces Apoptosis in Human Hepatoma Cells Through Activating 67-kDa Laminin Receptor/cGMP Signaling
Abstract
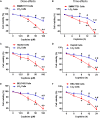
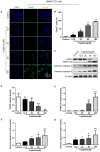
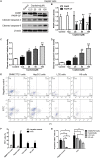
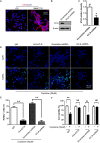
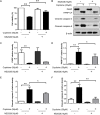
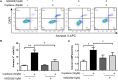
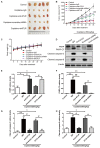
Hepatocellular carcinoma (HCC) is the most common primary cancer of the liver. Hence, new anti-liver cancer treatment strategies need to be urgently developed. Coptisine is a natural alkaloid extracted from rhizoma coptidis which exhibits anticancer activity in various preclinical models, including liver cancer. However, the molecular mechanisms underlying the anti-liver cancer effects of coptisine remains unclear. We used flow cytometry to assess the binding of coptisine to 67LR expressed on the surface of SMMC7721, HepG2, LO2 and H9 cells. Then SMMC7721, HepG2 and BEL7402 cells, belonging to the HCC cell lines, were treated with coptisine. The cell viability was detected using a cell counting kit-8 assay. Apoptosis was evaluated using flow cytometry and transferase-mediated dUTP nick-end labeling (TUNEL) assay. Apoptotic-related proteins and tumor death receptor 67-kDa laminin receptor (67LR) were detected using Western blot analysis. The cyclic guanosine 3',5'-monophosphate (cGMP) concentration was determined using enzyme-linked immunosorbent assay. sh67LR lentivirus, anti67LR antibody, and cGMP inhibitor NS2028 were used to determine how a 67LR/cGMP signaling pathway regulated coptisine-induced apoptosis. Tumor growth inhibited by coptisine was confirmed in a SMMC7721 cell xenograft mouse model. Coptisine selectively exhibited cell viability in human hepatoma cells but not in normal human hepatocyte cell line LO2 cells. Coptisine promoted SMMC7721 and HepG2 cell apoptosis by increasing 67LR activity. Both 67LR antibody and sh67LR treatment blocked coptisine-induced apoptosis and inhibition of cell viability. Coptisine upregulated the expression of cGMP. Moreover, cGMP inhibitor NS2028 significantly decreased coptisine-induced apoptosis and inhibition of cell viability. In vivo experiments confirmed that coptisine could significantly suppress the tumor growth and induce apoptosis in SMMC7721 xenografts through a 67LR/cGMP pathway. Coptisine-mediated 67LR activation may be a new therapeutic strategy for treating hepatic malignancy.
Keywords: 67LR; apoptosis; cGMP; coptisine; hepatocellular carcinoma.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

