Characterization of the Olfactory Receptor OR10H1 in Human Urinary Bladder Cancer
- PMID: 29867524
- PMCID: PMC5964926
- DOI: 10.3389/fphys.2018.00456
Characterization of the Olfactory Receptor OR10H1 in Human Urinary Bladder Cancer
Abstract
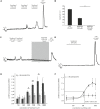
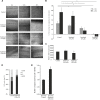
Olfactory receptors (ORs) are a large group of G-protein coupled receptors predominantly found in the olfactory epithelium. Many ORs are, however, ectopically expressed in other tissues and involved in several diseases including cancer. In this study, we describe that one OR, OR10H1, is predominantly expressed in the human urinary bladder with a notably higher expression at mRNA and protein level in bladder cancer tissues. Interestingly, also significantly higher amounts of OR10H1 transcripts were detectable in the urine of bladder cancer patients than in the urine of control persons. We identified the sandalwood-related compound Sandranol as a specific agonist of OR10H1. This deorphanization allowed the functional characterization of OR10H1 in BFTC905 bladder cancer cells. The effect of receptor activation was morphologically apparent in cell rounding, accompanied by changes in the cytoskeleton detected by β-actin, T-cadherin and β-Catenin staining. In addition, Sandranol treatment significantly diminished cell viability, cell proliferation and migration and induced a limited degree of apoptosis. Cell cycle analysis revealed an increased G1 fraction. In a concentration-dependent manner, Sandranol application elevated cAMP levels, which was reduced by inhibition of adenylyl cyclase, and elicited intracellular Ca2+ concentration increase. Furthermore, activation of OR10H1 enhanced secretion of ATP and serotonin. Our results suggest OR10H1 as a potential biomarker and therapeutic target for bladder cancer.
Keywords: OR10H1; biomarker; bladder; bladder cancer; next generation sequencing; olfactory receptor.
Figures





References
-
- Bommareddy A., Crisamore K., Fillman S., Brozena S., Steigerwalt J., Landis T., et al. (2015). Survivin Down-regulation by alpha-Santalol is not mediated through PI3K-AKT pathway in human breast cancer cells. 35 5353–5357. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

