The Use of a Combined Bioinformatics Approach to Locate Antibiotic Resistance Genes on Plasmids From Whole Genome Sequences of Salmonella enterica Serovars From Humans in Ghana
- PMID: 29867897
- PMCID: PMC5966558
- DOI: 10.3389/fmicb.2018.01010
The Use of a Combined Bioinformatics Approach to Locate Antibiotic Resistance Genes on Plasmids From Whole Genome Sequences of Salmonella enterica Serovars From Humans in Ghana
Abstract
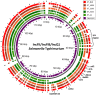
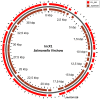
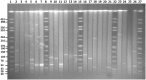
In the current study, we identified plasmids carrying antimicrobial resistance genes in draft whole genome sequences of 16 selected Salmonella enterica isolates representing six different serovars from humans in Ghana. The plasmids and the location of resistance genes in the genomes were predicted using a combination of PlasmidFinder, ResFinder, plasmidSPAdes and BLAST genomic analysis tools. Subsequently, S1-PFGE was employed for analysis of plasmid profiles. Whole genome sequencing confirmed the presence of antimicrobial resistance genes in Salmonella isolates showing multidrug resistance phenotypically. ESBL, either blaTEM52-B or blaCTX-M15 were present in two cephalosporin resistant isolates of S. Virchow and S. Poona, respectively. The systematic genome analysis revealed the presence of different plasmids in different serovars, with or without insertion of antimicrobial resistance genes. In S. Enteritidis, resistance genes were carried predominantly on plasmids of IncN type, in S. Typhimurium on plasmids of IncFII(S)/IncFIB(S)/IncQ1 type. In S. Virchow and in S. Poona, resistance genes were detected on plasmids of IncX1 and TrfA/IncHI2/IncHI2A type, respectively. The latter two plasmids were described for the first time in these serovars. The combination of genomic analytical tools allowed nearly full mapping of the resistance plasmids in all Salmonella strains analyzed. The results suggest that the improved analytical approach used in the current study may be used to identify plasmids that are specifically associated with resistance phenotypes in whole genome sequences. Such knowledge would allow the development of rapid multidrug resistance tracking tools in Salmonella populations using WGS.
Keywords: Ghana; Salmonella; multidrug resistance; plasmids; whole genome sequencing.
Figures



Similar articles
-
Genomic analyses reveal presence of extensively drug-resistant Salmonella enterica serovars isolated from clinical samples in Guizhou province, China, 2019-2023.Front Microbiol. 2025 Mar 27;16:1532036. doi: 10.3389/fmicb.2025.1532036. eCollection 2025. Front Microbiol. 2025. PMID: 40226105 Free PMC article.
-
Whole-Genome Sequence Analysis of an Extensively Drug-Resistant Salmonella enterica Serovar Agona Isolate from an Australian Silver Gull (Chroicocephalus novaehollandiae) Reveals the Acquisition of Multidrug Resistance Plasmids.mSphere. 2020 Nov 25;5(6):e00743-20. doi: 10.1128/mSphere.00743-20. mSphere. 2020. PMID: 33239365 Free PMC article.
-
Genome-Based Assessment of Antimicrobial Resistance and Virulence Potential of Isolates of Non-Pullorum/Gallinarum Salmonella Serovars Recovered from Dead Poultry in China.Microbiol Spectr. 2022 Aug 31;10(4):e0096522. doi: 10.1128/spectrum.00965-22. Epub 2022 Jun 21. Microbiol Spectr. 2022. PMID: 35727054 Free PMC article.
-
Genomic investigation of antimicrobial resistance determinants and virulence factors in Salmonella enterica serovars isolated from contaminated food and human stool samples in Brazil.Int J Food Microbiol. 2021 Apr 2;343:109091. doi: 10.1016/j.ijfoodmicro.2021.109091. Epub 2021 Feb 11. Int J Food Microbiol. 2021. PMID: 33639477
-
A Food Poisoning Caused by Salmonella Enterica (S. Enteritidis) ST11 Carrying Multi-Antimicrobial Resistance Genes in 2019, China.Infect Drug Resist. 2024 May 6;17:1751-1762. doi: 10.2147/IDR.S452295. eCollection 2024. Infect Drug Resist. 2024. PMID: 38736437 Free PMC article.
Cited by
-
Surveillance and Genomics of Toxigenic Vibrio cholerae O1 From Fish, Phytoplankton and Water in Lake Victoria, Tanzania.Front Microbiol. 2019 Apr 30;10:901. doi: 10.3389/fmicb.2019.00901. eCollection 2019. Front Microbiol. 2019. PMID: 31114556 Free PMC article.
-
Phenotypic and genotypic antimicrobial resistance correlation and plasmid characterization in Salmonella spp. isolates from Italy reveal high heterogeneity among serovars.Front Public Health. 2023 Sep 7;11:1221351. doi: 10.3389/fpubh.2023.1221351. eCollection 2023. Front Public Health. 2023. PMID: 37744490 Free PMC article.
-
The Plasmidomic Landscape of Clinical Methicillin-Resistant Staphylococcus aureus Isolates from Malaysia.Antibiotics (Basel). 2023 Apr 9;12(4):733. doi: 10.3390/antibiotics12040733. Antibiotics (Basel). 2023. PMID: 37107095 Free PMC article.
-
Antibiotic resistance: bioinformatics-based understanding as a functional strategy for drug design.RSC Adv. 2020 May 14;10(31):18451-18468. doi: 10.1039/d0ra01484b. eCollection 2020 May 10. RSC Adv. 2020. PMID: 35685616 Free PMC article. Review.
-
ESBL and AmpC β-Lactamase Encoding Genes in E. coli From Pig and Pig Farm Workers in Vietnam and Their Association With Mobile Genetic Elements.Front Microbiol. 2021 Mar 11;12:629139. doi: 10.3389/fmicb.2021.629139. eCollection 2021. Front Microbiol. 2021. PMID: 33776959 Free PMC article.
References
-
- Akinyemi K. O., Iwalokun B. A., Agboyinu J. A., Ogunyemi O., Fasure A. K. (2014). Emergence of third generation cephalosporin resistance and typing by Randomly amplified polymorphic DNA (RAPD) among clinical Salmonella isolates from Lagos, Nigeria. Br. Microbiol. Res. J. 4, 668–677. 10.9734/BMRJ/2014/8507 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

