Evaluating the Genetics of Common Variable Immunodeficiency: Monogenetic Model and Beyond
- PMID: 29867916
- PMCID: PMC5960686
- DOI: 10.3389/fimmu.2018.00636
Evaluating the Genetics of Common Variable Immunodeficiency: Monogenetic Model and Beyond
Abstract
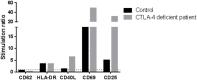
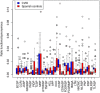
Common variable immunodeficiency (CVID) is the most frequent symptomatic primary immunodeficiency characterized by recurrent infections, hypogammaglobulinemia and poor response to vaccines. Its diagnosis is made based on clinical and immunological criteria, after exclusion of other diseases that can cause similar phenotypes. Currently, less than 20% of cases of CVID have a known underlying genetic cause. We have analyzed whole-exome sequencing and copy number variants data of 36 children and adolescents diagnosed with CVID and healthy relatives to estimate the proportion of monogenic cases. We have replicated an association of CVID to p.C104R in TNFRSF13B and reported the second case of homozygous patient to date. Our results also identify five causative genetic variants in LRBA, CTLA4, NFKB1, and PIK3R1, as well as other very likely causative variants in PRKCD, MAPK8, or DOCK8 among others. We experimentally validate the effect of the LRBA stop-gain mutation which abolishes protein production and downregulates the expression of CTLA4, and of the frameshift indel in CTLA4 producing expression downregulation of the protein. Our results indicate a monogenic origin of at least 15-24% of the CVID cases included in the study. The proportion of monogenic patients seems to be lower in CVID than in other PID that have also been analyzed by whole exome or targeted gene panels sequencing. Regardless of the exact proportion of CVID monogenic cases, other genetic models have to be considered for CVID. We propose that because of its prevalence and other features as intermediate penetrancies and phenotypic variation within families, CVID could fit with other more complex genetic scenarios. In particular, in this work, we explore the possibility of CVID being originated by an oligogenic model with the presence of heterozygous mutations in interacting proteins or by the accumulation of detrimental variants in particular immunological pathways, as well as perform association tests to detect association with rare genetic functional variation in the CVID cohort compared to healthy controls.
Keywords: common variable immunodeficiency; exome sequencing; loss-of-function; primary immunodeficiency; rare disease genetics.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

