STAT3-Mediated Transcriptional Regulation of Osteopontin in STAT3 Loss-of-Function Related Hyper IgE Syndrome
- PMID: 29868029
- PMCID: PMC5966547
- DOI: 10.3389/fimmu.2018.01080
STAT3-Mediated Transcriptional Regulation of Osteopontin in STAT3 Loss-of-Function Related Hyper IgE Syndrome
Abstract
Background: Hyper-IgE syndrome (HIES) caused by loss-of-function (LOF) mutations in STAT3 gene (STAT3 LOF HIES) is associated with dental and facial abnormalities in addition to immunological defects. The role of STAT3 in the pathogenesis of the dental/facial features is, however, poorly elucidated.
Objectives: Since mechanism of cellular resorption of mineralized tissues such as bone and teeth are similar, we attempted to study the expression of genes involved in bone homeostasis in STAT3 LOF HIES.
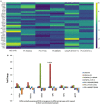
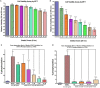
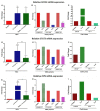
Methods: Peripheral blood mononuclear cells from healthy controls (HCs), STAT3 LOF HIES patients, STAT3-/- PC-3 cells and STAT3+/+ LNCaP cells were stimulated with IL-6 and quantitative PCR array was performed to study the relative mRNA expression of 43 pre-selected genes. PCR array finding were further evaluated after stattic induced STAT3 inhibition.
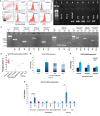
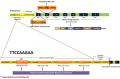
Results: Osteopontin (OPN) gene was seen to be significantly upregulated after IL-6 stimulation in HC (mean fold change 18.6, p = 0.01) compared with HIES subjects. Inhibition of STAT3 signaling by stattic followed by IL-6 stimulation abrogated the OPN response in HCs suggesting that IL-6-induced STAT3 signaling regulates OPN expression. Bioinformatics analysis predicted the presence of STAT3 response element TTCCAAGAA at position -2005 of the OPN gene.
Conclusion: Regulation of OPN gene through IL-6-mediated STAT3 activation and its significant dysregulation in STAT3 LOF HIES subjects could make OPN a plausible candidate involved in the pathogenesis of dental/facial manifestations in HIES.
Keywords: STAT3 signaling; craniosynostosis; dental anomalies; hyper-IgE syndrome; osteopontin; skeletal anomalies.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

