Alternaria Brassicae Induces Systemic Jasmonate Responses in Arabidopsis Which Travel to Neighboring Plants via a Piriformsopora Indica Hyphal Network and Activate Abscisic Acid Responses
- PMID: 29868082
- PMCID: PMC5952412
- DOI: 10.3389/fpls.2018.00626
Alternaria Brassicae Induces Systemic Jasmonate Responses in Arabidopsis Which Travel to Neighboring Plants via a Piriformsopora Indica Hyphal Network and Activate Abscisic Acid Responses
Abstract
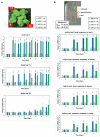
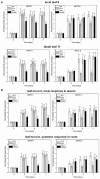
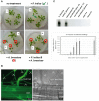
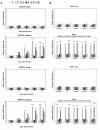
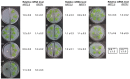
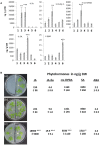
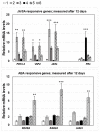
Stress information received by a particular local plant tissue is transferred to other tissues and neighboring plants, but how the information travels is not well understood. Application of Alternaria Brassicae spores to Arabidopsis leaves or roots stimulates local accumulation of jasmonic acid (JA), the expression of JA-responsive genes, as well as of NITRATE TRANSPORTER (NRT)2.5 and REDOX RESPONSIVE TRANSCRIPTION FACTOR1 (RRTF1). Infection information is systemically spread over the entire seedling and propagates radially from infected to non-infected leaves, axially from leaves to roots, and vice versa. The local and systemic NRT2.5 responses are reduced in the jar1 mutant, and the RRTF1 response in the rbohD mutant. Information about A. brassicae infection travels slowly to uninfected neighboring plants via a Piriformospora Indica hyphal network, where NRT2.5 and RRTF1 are up-regulated. The systemic A. brassicae-induced JA response in infected plants is converted to an abscisic acid (ABA) response in the neighboring plant where ABA and ABA-responsive genes are induced. We propose that the local threat information induced by A. brassicae infection is spread over the entire plant and transferred to neighboring plants via a P. indica hyphal network. The JA-specific response is converted to a general ABA-mediated stress response in the neighboring plant.
Keywords: Alternaria brassicae; NITRATE TRANSPORTER2.5; Piriformospora indica; REDOX RESPONSIVE TRANSCRIPTION FACTOR1; abscisic acid; interplant communication; jasmonic acid; systemic signaling.
Figures
Similar articles
-
High REDOX RESPONSIVE TRANSCRIPTION FACTOR1 Levels Result in Accumulation of Reactive Oxygen Species in Arabidopsis thaliana Shoots and Roots.Mol Plant. 2015 Aug;8(8):1253-73. doi: 10.1016/j.molp.2015.03.011. Epub 2015 Apr 13. Mol Plant. 2015. PMID: 25882345
-
Both the Jasmonic Acid and the Salicylic Acid Pathways Contribute to Resistance to the Biotrophic Clubroot Agent Plasmodiophora brassicae in Arabidopsis.Plant Cell Physiol. 2015 Nov;56(11):2158-68. doi: 10.1093/pcp/pcv127. Epub 2015 Sep 11. Plant Cell Physiol. 2015. PMID: 26363358
-
Jasmonic acid interacts with abscisic acid to regulate plant responses to water stress conditions.Plant Signal Behav. 2015;10(12):e1078953. doi: 10.1080/15592324.2015.1078953. Plant Signal Behav. 2015. PMID: 26340066 Free PMC article.
-
Onset of herbivore-induced resistance in systemic tissue primed for jasmonate-dependent defenses is activated by abscisic acid.Front Plant Sci. 2013 Dec 30;4:539. doi: 10.3389/fpls.2013.00539. eCollection 2013. Front Plant Sci. 2013. PMID: 24416038 Free PMC article.
-
Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots.Plant Cell. 1997 Dec;9(12):2243-59. doi: 10.1105/tpc.9.12.2243. Plant Cell. 1997. PMID: 9437865 Free PMC article.
Cited by
-
Beneficial and Pathogenic Arabidopsis Root-Interacting Fungi Differently Affect Auxin Levels and Responsive Genes During Early Infection.Front Microbiol. 2019 Mar 12;10:380. doi: 10.3389/fmicb.2019.00380. eCollection 2019. Front Microbiol. 2019. PMID: 30915043 Free PMC article.
-
Role of Phytohormones in Piriformospora indica-Induced Growth Promotion and Stress Tolerance in Plants: More Questions Than Answers.Front Microbiol. 2018 Jul 31;9:1646. doi: 10.3389/fmicb.2018.01646. eCollection 2018. Front Microbiol. 2018. PMID: 30140257 Free PMC article. Review.
-
The Impact of Piriformospora indica on plant heat and drought tolerance.Front Plant Sci. 2024 Dec 6;15:1479561. doi: 10.3389/fpls.2024.1479561. eCollection 2024. Front Plant Sci. 2024. PMID: 39711589 Free PMC article.
-
Ethylene Response Factor109 Attunes Immunity, Photosynthesis, and Iron Homeostasis in Arabidopsis Leaves.Front Plant Sci. 2022 Mar 2;13:841366. doi: 10.3389/fpls.2022.841366. eCollection 2022. Front Plant Sci. 2022. PMID: 35310669 Free PMC article.
-
Is Endophytic Colonization of Host Plants a Method of Alleviating Drought Stress? Conceptualizing the Hidden World of Endophytes.Int J Mol Sci. 2022 Aug 16;23(16):9194. doi: 10.3390/ijms23169194. Int J Mol Sci. 2022. PMID: 36012460 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

