PolyA tracks, polybasic peptides, poly-translational hurdles
- PMID: 29869837
- PMCID: PMC6281860
- DOI: 10.1002/wrna.1486
PolyA tracks, polybasic peptides, poly-translational hurdles
Abstract
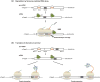
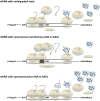
The abundance of messenger RNA (mRNA) is one of the major determinants of protein synthesis. As such, factors that influence mRNA stability often contribute to gene regulation. Polyadenylation of the 3' end of mRNA transcripts, the poly(A) tail, has long been recognized as one of these regulatory elements given its influence on translation efficiency and mRNA stability. Unwanted translation of the poly(A) tail signals to the cell an aberrant polyadenylation event or the lack of stop codons, which makes this sequence an important element in translation fidelity and mRNA surveillance response. Consequently, investigations into the effects of the poly(A) tail lead to the discoveries that poly-lysine as well as other polybasic peptide sequences and, to a much greater extent, polyA mRNA sequences within the open reading frame influence mRNA stability and translational efficiency. Conservation and evolutionary selection of codon usage in polyA track sequences across multiple organisms suggests a biological significance for coding polyA tracks in the regulation of gene expression. Here, we discuss the cellular responses and consequences of coding polyA track translation and synthesis of polybasic peptides. This article is categorized under: Translation > Translation Mechanisms Translation > Translation Regulation RNA Turnover and Surveillance > Turnover/Surveillance Mechanisms.
Keywords: RNA biology; mRNA turnover; translation; translational control.
© 2018 Wiley Periodicals, Inc.
Figures

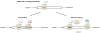
References
-
- Amaldi F, Pierandrei-Amaldi P. TO P Genes : A Translationally Controlled Class of Genes Including Those Coding for Ribosomal Proteins. Progress in Molecular and Subcellular Biology. 1997;18 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

