Receptor-Like Cytoplasmic Kinases Directly Link Diverse Pattern Recognition Receptors to the Activation of Mitogen-Activated Protein Kinase Cascades in Arabidopsis
- PMID: 29871986
- PMCID: PMC6096590
- DOI: 10.1105/tpc.17.00981
Receptor-Like Cytoplasmic Kinases Directly Link Diverse Pattern Recognition Receptors to the Activation of Mitogen-Activated Protein Kinase Cascades in Arabidopsis
Abstract
Plants deploy numerous cell surface-localized pattern-recognition receptors (PRRs) to perceive host- and microbe-derived molecular patterns that are specifically released during infection and activate defense responses. The activation of the mitogen-activated protein kinases MPK3, MPK4, and MPK6 (MPK3/4/6) is a hallmark of immune system activation by all known PRRs and is crucial for establishing disease resistance. The MAP kinase kinase kinase (MAPKKK) MEKK1 controls MPK4 activation, but the MAPKKKs responsible for MPK3/6 activation downstream of diverse PRRs and how the perception of diverse molecular patterns leads to the activation of MAPKKKs remain elusive. Here, we show that two highly related MAPKKKs, MAPKKK3 and MAPKKK5, mediate MPK3/6 activation by at least four PRRs and confer resistance to bacterial and fungal pathogens in Arabidopsis thaliana The receptor-like cytoplasmic kinases VII (RLCK VII), which act downstream of PRRs, directly phosphorylate MAPKKK5 Ser-599, which is required for pattern-triggered MPK3/6 activation, defense gene expression, and disease resistance. Surprisingly, MPK6 further phosphorylates MAPKKK5 Ser-682 and Ser-692 to enhance MPK3/6 activation and disease resistance, pointing to a positive feedback mechanism. Finally, MEKK1 Ser-603 is phosphorylated by both RLCK VII and MPK4, which is required for pattern-triggered MPK4 activation. These findings illustrate central mechanisms by which multiple PRRs activate MAPK cascades and disease resistance.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures


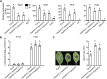
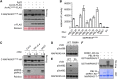
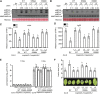

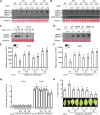


Comment in
-
RLCKs Bridge Plant Immune Receptors and MAPK Cascades.Trends Plant Sci. 2018 Dec;23(12):1039-1041. doi: 10.1016/j.tplants.2018.10.002. Epub 2018 Oct 17. Trends Plant Sci. 2018. PMID: 30342761
References
-
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. - PubMed
-
- Bayer M., Nawy T., Giglione C., Galli M., Meinnel T., Lukowitz W. (2009). Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323: 1485–1488. - PubMed
-
- Boller T., Felix G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60: 379–406. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

