The Tomato Hybrid Proline-rich Protein regulates the abscission zone competence to respond to ethylene signals
- PMID: 29872533
- PMCID: PMC5981600
- DOI: 10.1038/s41438-018-0033-2
The Tomato Hybrid Proline-rich Protein regulates the abscission zone competence to respond to ethylene signals
Abstract
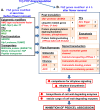
The Tomato Hybrid Proline-rich Protein (THyPRP) gene was specifically expressed in the tomato (Solanum lycopersicum) flower abscission zone (FAZ), and its stable antisense silencing under the control of an abscission zone (AZ)-specific promoter, Tomato Abscission Polygalacturonase4, significantly inhibited tomato pedicel abscission following flower removal. For understanding the THyPRP role in regulating pedicel abscission, a transcriptomic analysis of the FAZ of THyPRP-silenced plants was performed, using a newly developed AZ-specific tomato microarray chip. Decreased expression of THyPRP in the silenced plants was already observed before abscission induction, resulting in FAZ-specific altered gene expression of transcription factors, epigenetic modifiers, post-translational regulators, and transporters. Our data demonstrate that the effect of THyPRP silencing on pedicel abscission was not mediated by its effect on auxin balance, but by decreased ethylene biosynthesis and response. Additionally, THyPRP silencing revealed new players, which were demonstrated for the first time to be involved in regulating pedicel abscission processes. These include: gibberellin perception, Ca2+-Calmodulin signaling, Serpins and Small Ubiquitin-related Modifier proteins involved in post-translational modifications, Synthaxin and SNARE-like proteins, which participate in exocytosis, a process necessary for cell separation. These changes, occurring in the silenced plants early after flower removal, inhibited and/or delayed the acquisition of the competence of the FAZ cells to respond to ethylene signaling. Our results suggest that THyPRP acts as a master regulator of flower abscission in tomato, predominantly by playing a role in the regulation of the FAZ cell competence to respond to ethylene signals.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
-
- Osborne DJ. Abscission. Crit. Rev. Plant. Sci. 1989;8:103–129. doi: 10.1080/07352688909382272. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

