Pathway crosstalk enables cells to interpret TGF-β duration
- PMID: 29872541
- PMCID: PMC5972147
- DOI: 10.1038/s41540-018-0060-5
Pathway crosstalk enables cells to interpret TGF-β duration
Abstract
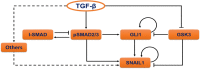
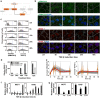
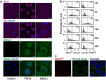
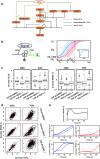
The detection and transmission of the temporal quality of intracellular and extracellular signals is an essential cellular mechanism. It remains largely unexplored how cells interpret the duration information of a stimulus. In this paper, we performed an integrated quantitative and computational analysis on TGF-β induced activation of SNAIL1, a key transcription factor that regulates several subsequent cell fate decisions such as apoptosis and epithelial-to-mesenchymal transition. We demonstrate that crosstalk among multiple TGF-β activated pathways forms a relay from SMAD to GLI1 that initializes and maintains SNAILl expression, respectively. SNAIL1 functions as a key integrator of information from TGF-β signaling distributed through upstream divergent pathways. The intertwined network serves as a temporal checkpoint, so that cells can generate a transient or sustained expression of SNAIL1 depending on TGF-β duration. Furthermore, we observed that TGF-β treatment leads to an unexpected accumulation of GSK3 molecules in an enzymatically active tyrosine phosphorylation form in Golgi apparatus and ER, followed by accumulation of GSK3 molecules in an enzymatically inhibitive serine phosphorylation in the nucleus. Subsequent model analysis and inhibition experiments revealed that the initial localized increase of GSK3 enzymatic activity couples to the positive feedback loop of the substrate Gli1 to form a network motif with multi-objective functions. That is, the motif is robust against stochastic fluctuations, and has a narrow distribution of response time that is insensitive to initial conditions. Specifically for TGF-β signaling, the motif ensures a smooth relay from SMAD to GLI1 on regulating SNAIL1 expression.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

