Systems medicine disease maps: community-driven comprehensive representation of disease mechanisms
- PMID: 29872544
- PMCID: PMC5984630
- DOI: 10.1038/s41540-018-0059-y
Systems medicine disease maps: community-driven comprehensive representation of disease mechanisms
Erratum in
-
Author Correction: Systems medicine disease maps: community-driven comprehensive representation of disease mechanisms.NPJ Syst Biol Appl. 2025 Jul 12;11(1):76. doi: 10.1038/s41540-025-00554-6. NPJ Syst Biol Appl. 2025. PMID: 40651992 Free PMC article. No abstract available.
Abstract
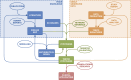
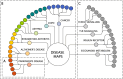
The development of computational approaches in systems biology has reached a state of maturity that allows their transition to systems medicine. Despite this progress, intuitive visualisation and context-dependent knowledge representation still present a major bottleneck. In this paper, we describe the Disease Maps Project, an effort towards a community-driven computationally readable comprehensive representation of disease mechanisms. We outline the key principles and the framework required for the success of this initiative, including use of best practices, standards and protocols. We apply a modular approach to ensure efficient sharing and reuse of resources for projects dedicated to specific diseases. Community-wide use of disease maps will accelerate the conduct of biomedical research and lead to new disease ontologies defined from mechanism-based disease endotypes rather than phenotypes.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Friend, S. H. & Norman, T. C. Metcalfe’s law and the biology information commons. Nat. Biotechnol.31, 297–303 (2013). - PubMed
-
- Dagley, S. & Nicholson D. E. An Introduction to Metabolic Pathways (Wiley, New York, 1970).
-
- Michal, G. (ed) Biochemical Pathways (wall chart) (Boehringer Mannheim, Mannheim, 1984).
LinkOut - more resources
Full Text Sources
Other Literature Sources

