A quantitative characterization of interaction between prion protein with nucleic acids
- PMID: 29872743
- PMCID: PMC5986701
- DOI: 10.1016/j.bbrep.2018.04.006
A quantitative characterization of interaction between prion protein with nucleic acids
Abstract
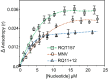
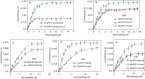
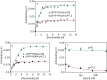
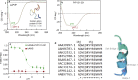
Binding of recombinant prion protein with small highly structured RNAs, prokaryotic and eukaryotic prion protein mRNA pseudoknots, tRNA and polyA has been studied by the change in fluorescence anisotropy of the intrinsic tryptophan groups of the protein. The affinities of these RNAs to the prion protein and the number of sites where the protein binds to the nucleic acids do not vary appreciably although the RNAs have very different compositions and structures. The binding parameters do not depend upon pH of the solution and show a poor co-operativity. The reactants form larger nucleoprotein complexes at pH 5 compared to that at neutral pH. The electrostatic force between the protein and nucleic acids dominates the binding interaction at neutral pH. In contrast, nucleic acid interaction with the incipient nonpolar groups exposed from the structured region of the prion protein dominates the reaction at pH 5. Prion protein of a particular species forms larger complexes with prion protein mRNA pseudoknots of the same species. The structure of the pseudoknots and not their base sequences probably dominates their interaction with prion protein. Possibilities of the conversion of the prion protein to its infectious form in the cytoplasm by nucleic acids have been discussed.
Keywords: Binding constant (Kd); Fluorescence anisotropy (r); Poly A; Prion protein; Pseudoknots; Small highly structured RNAs (shsRNAs).
Figures
Similar articles
-
Nucleic acid induced unfolding of recombinant prion protein globular fragment is pH dependent.Protein Sci. 2014 Dec;23(12):1780-8. doi: 10.1002/pro.2573. Epub 2014 Oct 28. Protein Sci. 2014. PMID: 25271002 Free PMC article.
-
Pseudoknots in prion protein mRNAs confirmed by comparative sequence analysis and pattern searching.Nucleic Acids Res. 2001 Feb 1;29(3):753-8. doi: 10.1093/nar/29.3.753. Nucleic Acids Res. 2001. PMID: 11160898 Free PMC article.
-
Small, highly structured RNAs participate in the conversion of human recombinant PrP(Sen) to PrP(Res) in vitro.J Mol Biol. 2003 Sep 5;332(1):47-57. doi: 10.1016/s0022-2836(03)00919-7. J Mol Biol. 2003. PMID: 12946346
-
Experimental approaches to the interaction of the prion protein with nucleic acids and glycosaminoglycans: Modulators of the pathogenic conversion.Methods. 2011 Mar;53(3):306-17. doi: 10.1016/j.ymeth.2010.12.002. Epub 2010 Dec 8. Methods. 2011. PMID: 21145399 Review.
-
Conformational changes of prion protein and nucleic acid arising from their interaction and relation of the altered structures in causing prion disease.Mini Rev Med Chem. 2008 Jul;8(8):784-9. doi: 10.2174/138955708784912201. Mini Rev Med Chem. 2008. PMID: 18673134 Review.
Cited by
-
A Dominant-Negative Mutant of ANXA7 Impairs Calcium Signaling and Enhances the Proliferation of Prostate Cancer Cells by Downregulating the IP3 Receptor and the PI3K/mTOR Pathway.Int J Mol Sci. 2023 May 16;24(10):8818. doi: 10.3390/ijms24108818. Int J Mol Sci. 2023. PMID: 37240163 Free PMC article.
-
RNA modulates aggregation of the recombinant mammalian prion protein by direct interaction.Sci Rep. 2019 Aug 27;9(1):12406. doi: 10.1038/s41598-019-48883-x. Sci Rep. 2019. PMID: 31455808 Free PMC article.
-
Molecular recognition and structural plasticity in amyloid-nucleic acid complexes.J Struct Biol. 2025 Jul 14;217(3):108233. doi: 10.1016/j.jsb.2025.108233. Online ahead of print. J Struct Biol. 2025. PMID: 40669763 Free PMC article.
-
Melatonin: Regulation of Prion Protein Phase Separation in Cancer Multidrug Resistance.Molecules. 2022 Jan 21;27(3):705. doi: 10.3390/molecules27030705. Molecules. 2022. PMID: 35163973 Free PMC article. Review.
-
Ribosomal RNA Modulates Aggregation of the Podospora Prion Protein HET-s.Int J Mol Sci. 2020 Sep 1;21(17):6340. doi: 10.3390/ijms21176340. Int J Mol Sci. 2020. PMID: 32882892 Free PMC article.
References
-
- Caughey B. Interactions between prion protein isoforms: the kiss of death? Trends Biochem. Sci. 2001;26(4):235–242. - PubMed
-
- Rachidi W., Mange A., Senator A., Guiraud P., Riondel J., Benboubetra M. Prion infection impairs copper binding of cultured cells. J. Biol. Chem. 2003;278(17):14595–14598. - PubMed
-
- Brown D.R., Schmidt B., Kretzschmar H.A. Effects of copper on survival of prion protein knockout neurons and glia. J. Neurochem. 1998;70(4):1686–1693. - PubMed
-
- Mouillet-Richard S., Ermonval M., Chebassier C., Laplanche J.L., Lehmann S., Launay J.M. Signal transduction through prion protein. Science. 2000;289(5486):1925–1928. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

