Prevalence of Clonal Hematopoiesis Mutations in Tumor-Only Clinical Genomic Profiling of Solid Tumors
- PMID: 29872864
- PMCID: PMC6224316
- DOI: 10.1001/jamaoncol.2018.2297
Prevalence of Clonal Hematopoiesis Mutations in Tumor-Only Clinical Genomic Profiling of Solid Tumors
Erratum in
-
Missing Conflict of Interest Disclosure.JAMA Oncol. 2019 Jan 1;5(1):122. doi: 10.1001/jamaoncol.2018.5678. JAMA Oncol. 2019. PMID: 30422161 Free PMC article. No abstract available.
Abstract
Importance: Although clonal hematopoiesis (CH) is well described in aging healthy populations, few studies have addressed the practical clinical implications of these alterations in solid-tumor sequencing.
Objective: To identify and quantify CH-related mutations in patients with solid tumors using matched tumor-blood sequencing, and to establish the proportion that would be misattributed to the tumor based on tumor-only sequencing (unmatched analysis).
Design, setting, and participants: Retrospective analysis of samples from 17 469 patients with solid cancers who underwent prospective clinical sequencing of DNA isolated from tumor tissue and matched peripheral blood using the MSK-IMPACT assay between January 2014 and August 2017.
Main outcomes and measures: We identified the presence of CH-related mutations in each patient's blood leukocytes and quantified the fraction of DNA molecules harboring the mutation in the corresponding matched tumor sample.
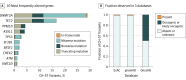
Results: The mean age of the 17 469 patients with cancer at sample collection was 59.2 years (range, 0.3-98.9 years); 53.6% were female. We identified 7608 CH-associated mutations in the blood of 4628 (26.5%) patients. A total of 1075 (14.1%) CH-associated mutations were also detectable in the matched tumor above established thresholds for calling somatic mutations. Overall, 912 (5.2%) patients would have had at least 1 CH-associated mutation erroneously called as tumor derived in the absence of matched blood sequencing. A total of 1061 (98.7%) of these mutations were absent from population scale databases of germline polymorphisms and therefore would have been challenging to filter informatically. Annotating variants with OncoKB classified 534 (49.7%) as oncogenic or likely oncogenic.
Conclusions and relevance: This study demonstrates how CH-derived mutations could lead to erroneous reporting and treatment recommendations when tumor-only sequencing is used.
Conflict of interest statement
Figures

Comment in
-
Failure to Accurately Disclose Conflicts of Interest in Articles Published in JAMA Oncology.JAMA Oncol. 2019 Jan 1;5(1):118-119. doi: 10.1001/jamaoncol.2018.5674. JAMA Oncol. 2019. PMID: 30422162 No abstract available.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

