Development, Validation, and Dissemination of a Breast Cancer Recurrence Detection and Timing Informatics Algorithm
- PMID: 29873757
- PMCID: PMC5972574
- DOI: 10.1093/jnci/djx200
Development, Validation, and Dissemination of a Breast Cancer Recurrence Detection and Timing Informatics Algorithm
Abstract
Background: This study developed, validated, and disseminated a generalizable informatics algorithm for detecting breast cancer recurrence and timing using a gold standard measure of recurrence coupled with data derived from a readily available common data model that pools health insurance claims and electronic health records data.
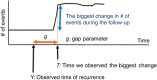
Methods: The algorithm has two parts: to detect the presence of recurrence and to estimate the timing of recurrence. The primary data source was the Cancer Research Network Virtual Data Warehouse (VDW). Sixteen potential indicators of recurrence were considered for model development. The final recurrence detection and timing models were determined, respectively, by maximizing the area under the ROC curve (AUROC) and minimizing average absolute error. Detection and timing algorithms were validated using VDW data in comparison with a gold standard recurrence capture from a third site in which recurrences were validated through chart review. Performance of this algorithm, stratified by stage at diagnosis, was compared with other published algorithms. All statistical tests were two-sided.
Results: Detection model AUROCs were 0.939 (95% confidence interval [CI] = 0.917 to 0.955) in the training data set (n = 3370) and 0.956 (95% CI = 0.944 to 0.971) and 0.900 (95% CI = 0.872 to 0.928), respectively, in the two validation data sets (n = 3370 and 3961, respectively). Timing models yielded average absolute prediction errors of 12.6% (95% CI = 10.5% to 14.5%) in the training data and 11.7% (95% CI = 9.9% to 13.5%) and 10.8% (95% CI = 9.6% to 12.2%) in the validation data sets, respectively, and were statistically significantly lower by 12.6% (95% CI = 8.8% to 16.5%, P < .001) than those estimated using previously reported timing algorithms. Similar covariates were included in both detection and timing algorithms but differed substantially from previous studies.
Conclusions: Valid and reliable detection of recurrence using data derived from electronic medical records and insurance claims is feasible. These tools will enable extensive, novel research on quality, effectiveness, and outcomes for breast cancer patients and those who develop recurrence.
Figures

Similar articles
-
Detecting Lung and Colorectal Cancer Recurrence Using Structured Clinical/Administrative Data to Enable Outcomes Research and Population Health Management.Med Care. 2017 Dec;55(12):e88-e98. doi: 10.1097/MLR.0000000000000404. Med Care. 2017. PMID: 29135771 Free PMC article.
-
Determining the Time of Cancer Recurrence Using Claims or Electronic Medical Record Data.JCO Clin Cancer Inform. 2018 Dec;2:1-10. doi: 10.1200/CCI.17.00163. JCO Clin Cancer Inform. 2018. PMID: 30652573 Free PMC article.
-
Performance of Cancer Recurrence Algorithms After Coding Scheme Switch From International Classification of Diseases 9th Revision to International Classification of Diseases 10th Revision.JCO Clin Cancer Inform. 2019 Mar;3:1-9. doi: 10.1200/CCI.18.00113. JCO Clin Cancer Inform. 2019. PMID: 30869998 Free PMC article.
-
A validated algorithm for register-based identification of patients with recurrence of breast cancer-Based on Danish Breast Cancer Group (DBCG) data.Cancer Epidemiol. 2019 Apr;59:129-134. doi: 10.1016/j.canep.2019.01.016. Epub 2019 Feb 8. Cancer Epidemiol. 2019. PMID: 30743224
-
A Systematic Review of Estimating Breast Cancer Recurrence at the Population Level With Administrative Data.J Natl Cancer Inst. 2020 Oct 1;112(10):979-988. doi: 10.1093/jnci/djaa050. J Natl Cancer Inst. 2020. PMID: 32259259 Free PMC article.
Cited by
-
Regarding the Utility of Unstructured Data and Natural Language Processing for Identification of Breast Cancer Recurrence.JCO Clin Cancer Inform. 2021 Sep;5:1024-1025. doi: 10.1200/CCI.21.00091. JCO Clin Cancer Inform. 2021. PMID: 34637320 Free PMC article. No abstract available.
-
Spending for Advanced Cancer Diagnoses: Comparing Recurrent Versus De Novo Stage IV Disease.J Oncol Pract. 2019 Jul;15(7):e616-e627. doi: 10.1200/JOP.19.00004. Epub 2019 May 20. J Oncol Pract. 2019. PMID: 31107629 Free PMC article.
-
Identification of recurrences in women diagnosed with early invasive breast cancer using routinely collected data in England.BJC Rep. 2025 May 28;3(1):39. doi: 10.1038/s44276-025-00154-1. BJC Rep. 2025. PMID: 40437042 Free PMC article.
-
New method for determining breast cancer recurrence-free survival using routinely collected real-world health data.BMC Cancer. 2022 Mar 16;22(1):281. doi: 10.1186/s12885-022-09333-6. BMC Cancer. 2022. PMID: 35296284 Free PMC article.
-
Utilization of the Cancer Medications Enquiry Database (CanMED)-National Drug Codes (NDC): Assessment of Systemic Breast Cancer Treatment Patterns.J Natl Cancer Inst Monogr. 2020 May 1;2020(55):46-52. doi: 10.1093/jncimonographs/lgaa002. J Natl Cancer Inst Monogr. 2020. PMID: 32412077 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

