Identification of a novel TIF-IA-NF-κB nucleolar stress response pathway
- PMID: 29873780
- PMCID: PMC6158704
- DOI: 10.1093/nar/gky455
Identification of a novel TIF-IA-NF-κB nucleolar stress response pathway
Abstract
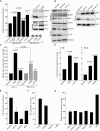
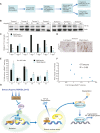
p53 as an effector of nucleolar stress is well defined, but p53 independent mechanisms are largely unknown. Like p53, the NF-κB transcription factor plays a critical role in maintaining cellular homeostasis under stress. Many stresses that stimulate NF-κB also disrupt nucleoli. However, the link between nucleolar function and activation of the NF-κB pathway is as yet unknown. Here we demonstrate that artificial disruption of the PolI complex stimulates NF-κB signalling. Unlike p53 nucleolar stress response, this effect does not appear to be linked to inhibition of rDNA transcription. We show that specific stress stimuli of NF-κB induce degradation of a critical component of the PolI complex, TIF-IA. This degradation precedes activation of NF-κB and is associated with increased nucleolar size. It is mimicked by CDK4 inhibition and is dependent upon a novel pathway involving UBF/p14ARF and S44 of the protein. We show that blocking TIF-IA degradation blocks stress effects on nucleolar size and NF-κB signalling. Finally, using ex vivo culture, we show a strong correlation between degradation of TIF-IA and activation of NF-κB in freshly resected, human colorectal tumours exposed to the chemopreventative agent, aspirin. Together, our study provides compelling evidence for a new, TIF-IA-NF-κB nucleolar stress response pathway that has in vivo relevance and therapeutic implications.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

