Single-Molecule Resolution of Antimicrobial Peptide Interactions with Supported Lipid A Bilayers
- PMID: 29874611
- PMCID: PMC6129183
- DOI: 10.1016/j.bpj.2018.04.019
Single-Molecule Resolution of Antimicrobial Peptide Interactions with Supported Lipid A Bilayers
Abstract
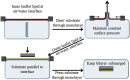
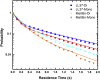
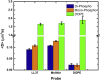
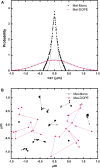
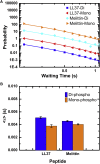
The molecular interactions between antimicrobial peptides (AMPs) and lipid A-containing supported lipid bilayers were probed using single-molecule total internal reflection fluorescence microscopy. Hybrid supported lipid bilayers with lipid A outer leaflets and phospholipid (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE)) inner leaflets were prepared and characterized, and the spatiotemporal trajectories of individual fluorescently labeled LL37 and Melittin AMPs were determined as they interacted with the bilayer surfaces comprising either monophosphoryl or diphosphoryl lipid A (from Escherichia coli) to determine the impact of electrostatic interactions. Large numbers of trajectories were obtained and analyzed to obtain the distributions of surface residence times and the statistics of the spatial trajectories. Interestingly, the AMP species were sensitive to subtle differences in the charge of the lipid, with both peptides diffusing more slowly and residing longer on the diphosphoryl lipid A. Furthermore, the single-molecule dynamics indicated a qualitative difference between the behavior of AMPs on hybrid Lipid A bilayers and on those composed entirely of DOPE. Whereas AMPs interacting with a DOPE bilayer exhibited two-dimensional Brownian diffusion with a diffusion coefficient of ∼1.7 μm2/s, AMPs adsorbed to the lipid A surface exhibited much slower apparent diffusion (on the order of ∼0.1 μm2/s) and executed intermittent trajectories that alternated between two-dimensional Brownian diffusion and desorption-mediated three-dimensional flights. Overall, these findings suggested that bilayers with lipid A in the outer leaflet, as it is in bacterial outer membranes, are valuable model systems for the study of the initial stage of AMP-bacterium interactions. Furthermore, single-molecule dynamics was sensitive to subtle differences in electrostatic interactions between cationic AMPs and monovalent or divalent anionic lipid A moieties.
Copyright © 2018 Biophysical Society. All rights reserved.
Figures

Similar articles
-
Mechanistic predictions of the influence of collagen-binding domain sequences on human LL37 interactions with model lipids using quartz crystal microbalance with dissipation.Biointerphases. 2019 Apr 30;14(2):021006. doi: 10.1116/1.5089759. Biointerphases. 2019. PMID: 31039613
-
Lateral diffusion of peripheral membrane proteins on supported lipid bilayers is controlled by the additive frictional drags of (1) bound lipids and (2) protein domains penetrating into the bilayer hydrocarbon core.Chem Phys Lipids. 2013 Jul-Aug;172-173:67-77. doi: 10.1016/j.chemphyslip.2013.04.005. Epub 2013 May 20. Chem Phys Lipids. 2013. PMID: 23701821 Free PMC article.
-
Membrane interaction of antimicrobial peptides using E. coli lipid extract as model bacterial cell membranes and SFG spectroscopy.Chem Phys Lipids. 2015 Apr;187:20-33. doi: 10.1016/j.chemphyslip.2015.02.003. Epub 2015 Feb 20. Chem Phys Lipids. 2015. PMID: 25707312 Free PMC article.
-
SFG studies on interactions between antimicrobial peptides and supported lipid bilayers.Biochim Biophys Acta. 2006 Sep;1758(9):1257-73. doi: 10.1016/j.bbamem.2006.01.017. Epub 2006 Feb 17. Biochim Biophys Acta. 2006. PMID: 16524559 Review.
-
Elementary Processes and Mechanisms of Interactions of Antimicrobial Peptides with Membranes-Single Giant Unilamellar Vesicle Studies.Adv Exp Med Biol. 2019;1117:17-32. doi: 10.1007/978-981-13-3588-4_3. Adv Exp Med Biol. 2019. PMID: 30980351 Review.
Cited by
-
Single-molecule phospholipase A2 becomes processive on melittin-induced membrane deformations.Biophys J. 2022 Apr 19;121(8):1417-1423. doi: 10.1016/j.bpj.2022.03.019. Epub 2022 Mar 18. Biophys J. 2022. PMID: 35314142 Free PMC article.
-
Antimicrobial peptide activity is anticorrelated with lipid a leaflet affinity.PLoS One. 2020 Nov 30;15(11):e0242907. doi: 10.1371/journal.pone.0242907. eCollection 2020. PLoS One. 2020. PMID: 33253275 Free PMC article.
-
Incorporation of Non-Canonical Amino Acids into Antimicrobial Peptides: Advances, Challenges, and Perspectives.Appl Environ Microbiol. 2022 Dec 13;88(23):e0161722. doi: 10.1128/aem.01617-22. Epub 2022 Nov 23. Appl Environ Microbiol. 2022. PMID: 36416555 Free PMC article. Review.
-
Membrane Active Peptides and Their Biophysical Characterization.Biomolecules. 2018 Aug 22;8(3):77. doi: 10.3390/biom8030077. Biomolecules. 2018. PMID: 30135402 Free PMC article. Review.
-
A Molecular Dynamics Study of Antimicrobial Peptide Interactions with the Lipopolysaccharides of the Outer Bacterial Membrane.J Membr Biol. 2022 Dec;255(6):665-675. doi: 10.1007/s00232-022-00258-6. Epub 2022 Aug 12. J Membr Biol. 2022. PMID: 35960325
References
-
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. - PubMed
-
- Pasupuleti M., Schmidtchen A., Malmsten M. Antimicrobial peptides: key components of the innate immune system. Crit. Rev. Biotechnol. 2012;32:143–171. - PubMed
-
- Christensen B., Fink J., Mauzerall D. Channel-forming properties of cecropins and related model compounds incorporated into planar lipid membranes (insect immunity/antibacterial peptide/voltage-dependent ion channel/bilayer lipid membrane/structure-function relation) Biophysics (Oxf.) 1988;85:5072–5076. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

