Control of Transmembrane Helix Dynamics by Interfacial Tryptophan Residues
- PMID: 29874612
- PMCID: PMC6129553
- DOI: 10.1016/j.bpj.2018.04.016
Control of Transmembrane Helix Dynamics by Interfacial Tryptophan Residues
Abstract
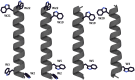
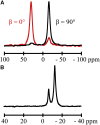
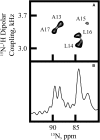
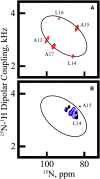
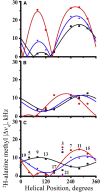
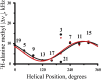
Transmembrane protein domains often contain interfacial aromatic residues, which may play a role in the insertion and stability of membrane helices. Residues such as Trp or Tyr, therefore, are often found situated at the lipid-water interface. We have examined the extent to which the precise radial locations of interfacial Trp residues may influence peptide helix orientation and dynamics. To address these questions, we have modified the GW5,19ALP23 (acetyl-GGALW5(LA)6LW19LAGA-[ethanol]amide) model peptide framework to relocate the Trp residues. Peptide orientation and dynamics were analyzed by means of solid-state nuclear magnetic resonance (NMR) spectroscopy to monitor specific 2H- and 15N-labeled residues. GW5,19ALP23 adopts a defined, tilted orientation within lipid bilayer membranes with minimal evidence of motional averaging of NMR observables, such as 2H quadrupolar or 15N-1H dipolar splittings. Here, we examine how peptide dynamics are impacted by relocating the interfacial Trp (W) residues on both ends and opposing faces of the helix, for example by a 100° rotation on the helical wheel for positions 4 and 20. In contrast to GW5,19ALP23, the modified GW4,20ALP23 helix experiences more extensive motional averaging of the NMR observables in several lipid bilayers of different thickness. Individual and combined Gaussian analyses of the 2H and 15N NMR signals confirm that the extent of dynamic averaging, particularly rotational "slippage" about the helix axis, is strongly coupled to the radial distribution of the interfacial Trp residues as well as the bilayer thickness. Additional 2H labels on alanines A3 and A21 reveal partial fraying of the helix ends. Even within the context of partial unwinding, the locations of particular Trp residues around the helix axis are prominent factors for determining transmembrane helix orientation and dynamics within the lipid membrane environment.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Breaking the Backbone: Central Arginine Residues Induce Membrane Exit and Helix Distortions within a Dynamic Membrane Peptide.J Phys Chem B. 2019 Sep 26;123(38):8034-8047. doi: 10.1021/acs.jpcb.9b06034. Epub 2019 Sep 17. J Phys Chem B. 2019. PMID: 31483653 Free PMC article.
-
Response of GWALP transmembrane peptides to changes in the tryptophan anchor positions.Biochemistry. 2011 Sep 6;50(35):7522-35. doi: 10.1021/bi2006459. Epub 2011 Aug 12. Biochemistry. 2011. PMID: 21800919 Free PMC article.
-
Comparisons of interfacial Phe, Tyr, and Trp residues as determinants of orientation and dynamics for GWALP transmembrane peptides.Biochemistry. 2014 Jun 10;53(22):3637-45. doi: 10.1021/bi500439x. Epub 2014 May 29. Biochemistry. 2014. PMID: 24829070 Free PMC article.
-
Solid-state NMR approaches to measure topological equilibria and dynamics of membrane polypeptides.Biochim Biophys Acta. 2010 Feb;1798(2):258-65. doi: 10.1016/j.bbamem.2009.06.021. Epub 2009 Jul 9. Biochim Biophys Acta. 2010. PMID: 19596252 Review.
-
Ca2+ -ATPase structure in the E1 and E2 conformations: mechanism, helix-helix and helix-lipid interactions.Biochim Biophys Acta. 2002 Oct 11;1565(2):246-66. doi: 10.1016/s0005-2736(02)00573-4. Biochim Biophys Acta. 2002. PMID: 12409199 Review.
Cited by
-
Phospholipid Membrane Interactions of Model Ac-WL-X-LL-OH Peptides Investigated by Solid-State Nuclear Magnetic Resonance.Membranes (Basel). 2024 May 1;14(5):105. doi: 10.3390/membranes14050105. Membranes (Basel). 2024. PMID: 38786939 Free PMC article.
-
Structural and Mechanismic Studies of Lactophoricin Analog, Novel Antibacterial Peptide.Int J Mol Sci. 2021 Apr 2;22(7):3734. doi: 10.3390/ijms22073734. Int J Mol Sci. 2021. PMID: 33918526 Free PMC article.
-
Probing the dynamic landscape of peptides in molecular assemblies by synergized NMR experiments and MD simulations.Commun Chem. 2024 Feb 13;7(1):28. doi: 10.1038/s42004-024-01115-4. Commun Chem. 2024. PMID: 38351219 Free PMC article.
-
Breaking the Backbone: Central Arginine Residues Induce Membrane Exit and Helix Distortions within a Dynamic Membrane Peptide.J Phys Chem B. 2019 Sep 26;123(38):8034-8047. doi: 10.1021/acs.jpcb.9b06034. Epub 2019 Sep 17. J Phys Chem B. 2019. PMID: 31483653 Free PMC article.
-
Lipid-Dependent Titration of Glutamic Acid at a Bilayer Membrane Interface.ACS Omega. 2021 Mar 17;6(12):8488-8494. doi: 10.1021/acsomega.1c00276. eCollection 2021 Mar 30. ACS Omega. 2021. PMID: 33817510 Free PMC article.
References
-
- Yau W.M., Wimley W.C., White S.H. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. - PubMed
-
- Koeppe R.E., II, Anderson O.S. Engineering the gramicidin channel. Annu. Rev. Biophys. Biomol. Struct. 1996;25:231–258. - PubMed
-
- O’Connell A.M., Koeppe R.E., II, Andersen O.S. Kinetics of gramicidin channel formation in lipid bilayers: transmembrane monomer association. Science. 1990;250:1256–1259. - PubMed
-
- McKay M.J., Afrose F., Greathouse D.V. Helix formation and stability in membranes. Biochim. Biophys. Acta. 2018 Published online February 13, 2018. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

