Emerging Technologies for Cancer Research: Towards Personalized Medicine with Microfluidic Platforms and 3D Tumor Models
- PMID: 29874987
- PMCID: PMC6302350
- DOI: 10.2174/0929867325666180605122633
Emerging Technologies for Cancer Research: Towards Personalized Medicine with Microfluidic Platforms and 3D Tumor Models
Abstract
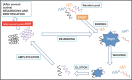
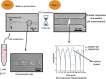
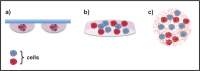
In the present review, we describe three hot topics in cancer research such as circulating tumor cells, exosomes, and 3D environment models. The first section is dedicated to microfluidic platforms for detecting circulating tumor cells, including both affinity-based methods that take advantage of antibodies and aptamers, and "label-free" approaches, exploiting cancer cells physical features and, more recently, abnormal cancer metabolism. In the second section, we briefly describe the biology of exosomes and their role in cancer, as well as conventional techniques for their isolation and innovative microfluidic platforms. In the third section, the importance of tumor microenvironment is highlighted, along with techniques for modeling it in vitro. Finally, we discuss limitations of two-dimensional monolayer methods and describe advantages and disadvantages of different three-dimensional tumor systems for cell-cell interaction analysis and their potential applications in cancer management.
Keywords: 3D cell cultures; Cancer; circulating tumor cells; exosomes; microfluidic Platforms; tumor microenvironment..
Copyright© Bentham Science Publishers; For any queries, please email at epub@benthamscience.org.
Figures




References
-
- Stewart B. W., Wild C. P. World Cancer Report 2014 . 2014.
-
- Li H., Fan X., Houghton J. Tumor microenvironment: The role of the tumor stroma in cancer. J. Cell. Biochem. 2007:805–815. - PubMed
-
- Wistuba I.I., Gelovani J.G., Jacoby J.J., Davis S.E., Herbst R.S. Methodological and practical challenges for personalized cancer therapies. Nat. Rev. Clin. Oncol. 2011;8(3):135–141. - PubMed
-
- Schilsky R.L. Personalized medicine in oncology: The future is now. Nat. Rev. Drug Discov. 2010;9:363–366. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

