Mamu-B*17+ Rhesus Macaques Vaccinated with env, vif, and nef Manifest Early Control of SIVmac239 Replication
- PMID: 29875239
- PMCID: PMC6069176
- DOI: 10.1128/JVI.00690-18
Mamu-B*17+ Rhesus Macaques Vaccinated with env, vif, and nef Manifest Early Control of SIVmac239 Replication
Abstract
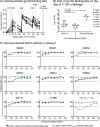
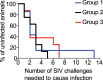
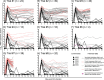
Certain major histocompatibility complex class I (MHC-I) alleles are associated with spontaneous control of viral replication in human immunodeficiency virus (HIV)-infected people and simian immunodeficiency virus (SIV)-infected rhesus macaques (RMs). These cases of "elite" control of HIV/SIV replication are often immune-mediated, thereby providing a framework for studying anti-lentiviral immunity. In this study, we examined how vaccination impacts SIV replication in RMs expressing the MHC-I allele Mamu-B*17 Approximately 21% of Mamu-B*17+ and 50% of Mamu-B*08+ RMs control chronic-phase viremia after SIVmac239 infection. Because CD8+ T cells targeting Mamu-B*08-restricted SIV epitopes have been implicated in virologic suppression in Mamu-B*08+ RMs, we investigated whether this might also be true for Mamu-B*17+ RMs. Two groups of Mamu-B*17+ RMs were vaccinated with genes encoding Mamu-B*17-restricted epitopes in Vif and Nef. These genes were delivered by themselves (group 1) or together with env (group 2). Group 3 included MHC-I-matched RMs and served as the control group. Surprisingly, the group 1 vaccine regimen had little effect on viral replication compared to group 3, suggesting that unlike Mamu-B*08+ RMs, preexisting SIV-specific CD8+ T cells alone do not facilitate long-term virologic suppression in Mamu-B*17+ RMs. Remarkably, however, 5/8 group 2 vaccinees controlled viremia to <15 viral RNA copies/ml soon after infection. No serological neutralizing activity against SIVmac239 was detected in group 2, although vaccine-elicited gp140-binding antibodies correlated inversely with nadir viral loads. Collectively, these data shed new light on the unique mechanism of elite control in Mamu-B*17+ RMs and implicate vaccine-induced, nonneutralizing anti-Env antibodies in the containment of immunodeficiency virus infection.IMPORTANCE A better understanding of the immune correlates of protection against HIV might facilitate the development of a prophylactic vaccine. Therefore, we investigated simian immunodeficiency virus (SIV) infection outcomes in rhesus macaques expressing the major histocompatibility complex class I allele Mamu-B*17 Approximately 21% of Mamu-B*17+ macaques spontaneously controlled chronic phase viremia after SIV infection, an effect that may involve CD8+ T cells targeting Mamu-B*17-restricted SIV epitopes. We vaccinated Mamu-B*17+ macaques with genes encoding immunodominant epitopes in Vif and Nef alone (group 1) or together with env (group 2). Although neither vaccine regimen prevented SIV infection, 5/8 group 2 vaccinees controlled viremia to below detection limits shortly after infection. This outcome, which was not observed in group 1, was associated with vaccine-induced, nonneutralizing Env-binding antibodies. Together, these findings suggest a limited contribution of Vif- and Nef-specific CD8+ T cells for virologic control in Mamu-B*17+ macaques and implicate anti-Env antibodies in containment of SIV infection.
Keywords: AIDS; human immunodeficiency virus; simian immunodeficiency virus; vaccines.
Copyright © 2018 American Society for Microbiology.
Figures
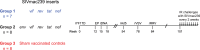








References
-
- UNAIDS. 2016. Global AIDS update. UNAIDS, Geneva, Switzerland.
-
- Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. - DOI - PMC - PubMed
-
- Gray GE, Allen M, Moodie Z, Churchyard G, Bekker LG, Nchabeleng M, Mlisana K, Metch B, de Bruyn G, Latka MH, Roux S, Mathebula M, Naicker N, Ducar C, Carter DK, Puren A, Eaton N, McElrath MJ, Robertson M, Corey L, Kublin JG. 2011. Safety and efficacy of the HVTN 503/Phambili study of a clade-B-based HIV-1 vaccine in South Africa: a double-blind, randomised, placebo-controlled test-of-concept phase 2b study. Lancet Infect Dis 11:507–515. doi: 10.1016/S1473-3099(11)70098-6. - DOI - PMC - PubMed
-
- Hammer SM, Sobieszczyk ME, Janes H, Karuna ST, Mulligan MJ, Grove D, Koblin BA, Buchbinder SP, Keefer MC, Tomaras GD, Frahm N, Hural J, Anude C, Graham BS, Enama ME, Adams E, DeJesus E, Novak RM, Frank I, Bentley C, Ramirez S, Fu R, Koup RA, Mascola JR, Nabel GJ, Montefiori DC, Kublin J, McElrath MJ, Corey L, Gilbert PB. 2013. Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N Engl J Med 369:2083–2092. doi: 10.1056/NEJMoa1310566. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI108421/AI/NIAID NIH HHS/United States
- R37 AI052056/AI/NIAID NIH HHS/United States
- R01 AI095098/AI/NIAID NIH HHS/United States
- R56 AI049120/AI/NIAID NIH HHS/United States
- K01 OD023032/OD/NIH HHS/United States
- R01 AI121135/AI/NIAID NIH HHS/United States
- R37 AI063928/AI/NIAID NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- R01 AI098485/AI/NIAID NIH HHS/United States
- P01 AI104715/AI/NIAID NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- P51 OD011106/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

