Phage-inducible chromosomal islands are ubiquitous within the bacterial universe
- PMID: 29875435
- PMCID: PMC6092414
- DOI: 10.1038/s41396-018-0156-3
Phage-inducible chromosomal islands are ubiquitous within the bacterial universe
Abstract
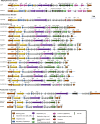
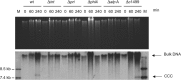
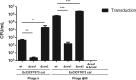
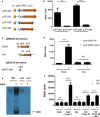
Phage-inducible chromosomal islands (PICIs) are a recently discovered family of pathogenicity islands that contribute substantively to horizontal gene transfer, host adaptation and virulence in Gram-positive cocci. Here we report that similar elements also occur widely in Gram-negative bacteria. As with the PICIs from Gram-positive cocci, their uniqueness is defined by a constellation of features: unique and specific attachment sites, exclusive PICI genes, a phage-dependent mechanism of induction, conserved replication origin organization, convergent mechanisms of phage interference, and specific packaging of PICI DNA into phage-like infectious particles, resulting in very high transfer frequencies. We suggest that the PICIs represent two or more distinct lineages, have spread widely throughout the bacterial world, and have diverged much more slowly than their host organisms or their prophage cousins. Overall, these findings represent the discovery of a universal class of mobile genetic elements.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Ubeda C, Tormo MA, Cucarella C, Trotonda P, Foster TJ, Lasa I, et al. Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol Microbiol. 2003;49:193–210. doi: 10.1046/j.1365-2958.2003.03577.x. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

