Autophagy and Its Interaction With Intracellular Bacterial Pathogens
- PMID: 29875765
- PMCID: PMC5974045
- DOI: 10.3389/fimmu.2018.00935
Autophagy and Its Interaction With Intracellular Bacterial Pathogens
Abstract
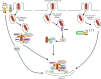
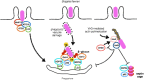
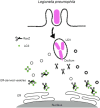
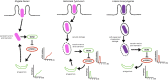
Cellular responses to stress can be defined by the overwhelming number of changes that cells go through upon contact with and stressful conditions such as infection and modifications in nutritional status. One of the main cellular responses to stress is autophagy. Much progress has been made in the understanding of the mechanisms involved in the induction of autophagy during infection by intracellular bacteria. This review aims to discuss recent findings on the role of autophagy as a cellular response to intracellular bacterial pathogens such as, Streptococcus pyogenes, Mycobacterium tuberculosis, Shigella flexneri, Salmonella typhimurium, Listeria monocytogenes, and Legionella pneumophila, how the autophagic machinery senses these bacteria directly or indirectly (through the detection of bacteria-induced nutritional stress), and how some of these bacterial pathogens manage to escape from autophagy.
Keywords: Legionella pneumophila; Listeria monocytogenes; Mycobacterium tuberculosis; Salmonella typhimurium; Shigella flexneri; Streptococcus pyogenes; autophagy; infection.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

