Genomic analyses identify multiple Asian origins and deeply diverged mitochondrial clades in inbred brown rats (Rattus norvegicus)
- PMID: 29875813
- PMCID: PMC5979757
- DOI: 10.1111/eva.12572
Genomic analyses identify multiple Asian origins and deeply diverged mitochondrial clades in inbred brown rats (Rattus norvegicus)
Abstract
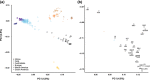
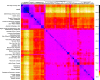
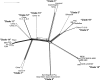
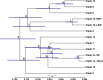
Over 500 strains of inbred brown rats (Rattus norvegicus) have been developed for use as a biomedical model organism. Most of these inbred lines were derived from the colony established at the Wistar Institute in 1906 or its descendants following worldwide distribution to research and breeding centers. The geographic source of the animals that founded the Wistar colony has been lost to history; thus, we compared 25 inbred rat strains to 326 wild rats from a global diversity dataset at 32 k SNPs, and 47 mitochondrial genomes to identify the source populations. We analyzed nuclear genomic data using principal component analyses and co-ancestry heat maps, and mitogenomes using phylogenetic trees and networks. In the nuclear genome, inbred rats clustered together indicating a single geographic origin for the strains studied and showed admixed ancestral variation with wild rats in eastern Asia and western North America. The Sprague Dawley derived, Wistar derived, and Brown Norway strains each had mitogenomes from different clades which diverged between 13 and 139 kya. Thus, we posit that rats originally collected for captive breeding had high mitochondrial diversity that became fixed through genetic drift and/or artificial selection. Our results show that these important medical models share common genomic ancestry from a few source populations, and opportunities exist to create new strains with diverse genomic backgrounds to provide novel insight into the genomic basis of disease phenotypes.
Keywords: inbreeding; mitogenomes; mitonuclear discordance; rat strains.
Figures
References
-
- Abhyankar, A. , Park, H.‐B. , Tonolo, G. , & Luthman, H. (2009). Comparative sequence analysis of the non‐protein‐coding mitochondrial DNA of inbred rat strains. PLoS One, 4(12), e8148 https://doi.org/10.1371/journal.pone.0008148 - DOI - PMC - PubMed
-
- Aitman, T. J. , Critser, J. K. , Cuppen, E. , Dominiczak, A. , Fernandez‐Suarez, X. M. , Flint, J. , … Worley, K. (2008). Progress and prospects in rat genetics: A community view. Nature Genetics, 40(5), 516–522. https://doi.org/10.1038/ng.147 - DOI - PubMed
-
- Atanur, S. S. , Diaz, A. G. , Maratou, K. , Sarkis, A. , Rotival, M. , Game, L. , … Aitman, T. J. (2013). Genome sequencing reveals loci under artificial selection that underlie disease phenotypes in the laboratory rat. Cell, 154(3), 691–703. https://doi.org/10.1016/j.cell.2013.06.040 - DOI - PMC - PubMed
-
- Balakirev, A. E. , & Rozhnov, V. V. (2012). Contribution to the species composition and taxonomic status of some Rattus inhabiting Southern Vietnam and Sundaland. Russian Journal of Theriology, 11(1), 33–45. https://doi.org/10.15298/rusjtheriol.11.1.03 - DOI
-
- Bastos, A. D. , Nair, D. , Taylor, P. J. , Brettschneider, H. , Kirsten, F. , Mostert, E. , … Chimimba, C. T. (2011). Genetic monitoring detects an overlooked cryptic species and reveals the diversity and distribution of three invasive Rattus congeners in South Africa. BMC Genetics, 12, 26 https://doi.org/10.1186/1471-2156-12-26 - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

