Addicted to sugar: roles of glycans in the order Mononegavirales
- PMID: 29878112
- PMCID: PMC6291800
- DOI: 10.1093/glycob/cwy053
Addicted to sugar: roles of glycans in the order Mononegavirales
Abstract
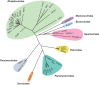
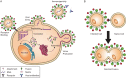
Glycosylation is a biologically important protein modification process by which a carbohydrate chain is enzymatically added to a protein at a specific amino acid residue. This process plays roles in many cellular functions, including intracellular trafficking, cell-cell signaling, protein folding and receptor binding. While glycosylation is a common host cell process, it is utilized by many pathogens as well. Protein glycosylation is widely employed by viruses for both host invasion and evasion of host immune responses. Thus better understanding of viral glycosylation functions has potential applications for improved antiviral therapeutic and vaccine development. Here, we summarize our current knowledge on the broad biological functions of glycans for the Mononegavirales, an order of enveloped negative-sense single-stranded RNA viruses of high medical importance that includes Ebola, rabies, measles and Nipah viruses. We discuss glycobiological findings by genera in alphabetical order within each of eight Mononegavirales families, namely, the bornaviruses, filoviruses, mymonaviruses, nyamiviruses, paramyxoviruses, pneumoviruses, rhabdoviruses and sunviruses.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

