QQS orphan gene and its interactor NF-YC4 reduce susceptibility to pathogens and pests
- PMID: 29878511
- PMCID: PMC6330549
- DOI: 10.1111/pbi.12961
QQS orphan gene and its interactor NF-YC4 reduce susceptibility to pathogens and pests
Abstract
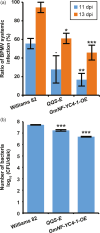
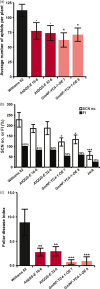
Enhancing the nutritional quality and disease resistance of crops without sacrificing productivity is a key issue for developing varieties that are valuable to farmers and for simultaneously improving food security and sustainability. Expression of the Arabidopsis thaliana species-specific AtQQS (Qua-Quine Starch) orphan gene or its interactor, NF-YC4 (Nuclear Factor Y, subunit C4), has been shown to increase levels of leaf/seed protein without affecting the growth and yield of agronomic species. Here, we demonstrate that overexpression of AtQQS and NF-YC4 in Arabidopsis and soybean enhances resistance/reduces susceptibility to viruses, bacteria, fungi, aphids and soybean cyst nematodes. A series of Arabidopsis mutants in starch metabolism were used to explore the relationships between QQS expression, carbon and nitrogen partitioning, and defense. The enhanced basal defenses mediated by QQS were independent of changes in protein/carbohydrate composition of the plants. We demonstrate that either AtQQS or NF-YC4 overexpression in Arabidopsis and in soybean reduces susceptibility of these plants to pathogens/pests. Transgenic soybean lines overexpressing NF-YC4 produce seeds with increased protein while maintaining healthy growth. Pull-down studies reveal that QQS interacts with human NF-YC, as well as with Arabidopsis NF-YC4, and indicate two QQS binding sites near the NF-YC-histone-binding domain. A new model for QQS interaction with NF-YC is speculated. Our findings illustrate the potential of QQS and NF-YC4 to increase protein and improve defensive traits in crops, overcoming the normal growth-defense trade-offs.
Keywords: QQS; NF-YC4; carbon and nitrogen partitioning; orphan; pathogen; pest.
© 2018 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures




References
-
- Arendsee, Z.W. , Li, L. and Wurtele, E.S. (2014) Coming of age: orphan genes in plants. Trends Plant Sci. 19, 698–708. - PubMed
-
- Badel, J.L. , Shimizu, R. , Oh, H.S. and Collmer, A. (2006) A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol. Plant Microbe Interact. 19, 99–111. - PubMed
-
- Blackman, R.L. and Eastop, V.F. (2000) Aphids on the World's Crops: An Identification and Information Guide. Chichester and New York, NY: Wiley.
-
- Bolton, M.D. (2009) Primary metabolism and plant defense–fuel for the fire. Mol. Plant Microbe Interact. 22, 487–497. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

