Predicting 3D structure and stability of RNA pseudoknots in monovalent and divalent ion solutions
- PMID: 29879103
- PMCID: PMC6007934
- DOI: 10.1371/journal.pcbi.1006222
Predicting 3D structure and stability of RNA pseudoknots in monovalent and divalent ion solutions
Abstract
RNA pseudoknots are a kind of minimal RNA tertiary structural motifs, and their three-dimensional (3D) structures and stability play essential roles in a variety of biological functions. Therefore, to predict 3D structures and stability of RNA pseudoknots is essential for understanding their functions. In the work, we employed our previously developed coarse-grained model with implicit salt to make extensive predictions and comprehensive analyses on the 3D structures and stability for RNA pseudoknots in monovalent/divalent ion solutions. The comparisons with available experimental data show that our model can successfully predict the 3D structures of RNA pseudoknots from their sequences, and can also make reliable predictions for the stability of RNA pseudoknots with different lengths and sequences over a wide range of monovalent/divalent ion concentrations. Furthermore, we made comprehensive analyses on the unfolding pathway for various RNA pseudoknots in ion solutions. Our analyses for extensive pseudokonts and the wide range of monovalent/divalent ion concentrations verify that the unfolding pathway of RNA pseudoknots is mainly dependent on the relative stability of unfolded intermediate states, and show that the unfolding pathway of RNA pseudoknots can be significantly modulated by their sequences and solution ion conditions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






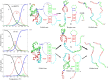
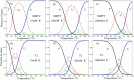
Similar articles
-
Predicting 3D structures and stabilities for complex RNA pseudoknots in ion solutions.Biophys J. 2023 Apr 18;122(8):1503-1516. doi: 10.1016/j.bpj.2023.03.017. Epub 2023 Mar 15. Biophys J. 2023. PMID: 36924021 Free PMC article.
-
Structure folding of RNA kissing complexes in salt solutions: predicting 3D structure, stability, and folding pathway.RNA. 2019 Nov;25(11):1532-1548. doi: 10.1261/rna.071662.119. Epub 2019 Aug 7. RNA. 2019. PMID: 31391217 Free PMC article.
-
Predicting 3D Structure, Flexibility, and Stability of RNA Hairpins in Monovalent and Divalent Ion Solutions.Biophys J. 2015 Dec 15;109(12):2654-2665. doi: 10.1016/j.bpj.2015.11.006. Biophys J. 2015. PMID: 26682822 Free PMC article.
-
Probing the kinetic and thermodynamic consequences of the tetraloop/tetraloop receptor monovalent ion-binding site in P4-P6 RNA by smFRET.Biochem Soc Trans. 2015 Apr;43(2):172-8. doi: 10.1042/BST20140268. Biochem Soc Trans. 2015. PMID: 25849913 Free PMC article. Review.
-
RNA pseudoknots and the regulation of protein synthesis.Biochem Soc Trans. 2008 Aug;36(Pt 4):684-9. doi: 10.1042/BST0360684. Biochem Soc Trans. 2008. PMID: 18631140 Review.
Cited by
-
Structural Flexibility of DNA-RNA Hybrid Duplex: Stretching and Twist-Stretch Coupling.Biophys J. 2019 Jul 9;117(1):74-86. doi: 10.1016/j.bpj.2019.05.018. Epub 2019 May 23. Biophys J. 2019. PMID: 31164196 Free PMC article.
-
ABC2A: A Straightforward and Fast Method for the Accurate Backmapping of RNA Coarse-Grained Models to All-Atom Structures.Molecules. 2024 Mar 11;29(6):1244. doi: 10.3390/molecules29061244. Molecules. 2024. PMID: 38542881 Free PMC article.
-
3dRNA: Building RNA 3D structure with improved template library.Comput Struct Biotechnol J. 2020 Aug 28;18:2416-2423. doi: 10.1016/j.csbj.2020.08.017. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 33005304 Free PMC article.
-
What is the best reference state for building statistical potentials in RNA 3D structure evaluation?RNA. 2019 Jul;25(7):793-812. doi: 10.1261/rna.069872.118. Epub 2019 Apr 17. RNA. 2019. PMID: 30996105 Free PMC article.
-
Salt-Dependent RNA Pseudoknot Stability: Effect of Spatial Confinement.Front Mol Biosci. 2021 Apr 13;8:666369. doi: 10.3389/fmolb.2021.666369. eCollection 2021. Front Mol Biosci. 2021. PMID: 33928126 Free PMC article.
References
-
- Atkins JF, Gesteland RF, Cech TR. RNA worlds: From life’s origins to diversity in gene regulation Cold Spring Harbor Laboratory Press Cold Spring Harbor, NY; 2011.
-
- Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: Structure and mechanism. Virus Res. 2009; 139: 193–208. doi: 10.1016/j.virusres.2008.06.008 - DOI - PMC - PubMed
-
- Su L, Chen L, Egli M, Berger JM. Rich A. Minor groove RNA triplex in the crystal structure of a ribosomal frameshifting viral pseudoknot. Nat. Struct. Biol. 1999; 6: 285–292. doi: 10.1038/6722 - DOI - PMC - PubMed
-
- Tinoco I, Bustamante C. How RNA folds. J. Mol. Biol. 1999; 293: 271–281. doi: 10.1006/jmbi.1999.3001 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

