METTL3-mediated m6A modification is required for cerebellar development
- PMID: 29879109
- PMCID: PMC6021109
- DOI: 10.1371/journal.pbio.2004880
METTL3-mediated m6A modification is required for cerebellar development
Abstract
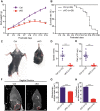
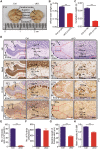
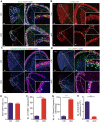
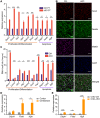
N6-methyladenosine (m6A) RNA methylation is the most abundant modification on mRNAs and plays important roles in various biological processes. The formation of m6A is catalyzed by a methyltransferase complex including methyltransferase-like 3 (METTL3) as a key factor. However, the in vivo functions of METTL3 and m6A modification in mammalian development remain unclear. Here, we show that specific inactivation of Mettl3 in mouse nervous system causes severe developmental defects in the brain. Mettl3 conditional knockout (cKO) mice manifest cerebellar hypoplasia caused by drastically enhanced apoptosis of newborn cerebellar granule cells (CGCs) in the external granular layer (EGL). METTL3 depletion-induced loss of m6A modification causes extended RNA half-lives and aberrant splicing events, consequently leading to dysregulation of transcriptome-wide gene expression and premature CGC death. Our findings reveal a critical role of METTL3-mediated m6A in regulating the development of mammalian cerebellum.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Hsieh J, Zhao X. Genetics and Epigenetics in Adult Neurogenesis. Cold Spring Harb Perspect Biol. 2016;8: a018911 doi: 10.1101/cshperspect.a018911 - DOI - PMC - PubMed
-
- Hwang JY, Aromolaran KA, Zukin RS. The emerging field of epigenetics in neurodegeneration and neuroprotection. Nat Rev Neurosci. 2017;18: 347–361. doi: 10.1038/nrn.2017.46 - DOI - PMC - PubMed
-
- Cholewa-Waclaw J, Bird A, von Schimmelmann M, Schaefer A, Yu H, Song H, et al. The Role of Epigenetic Mechanisms in the Regulation of Gene Expression in the Nervous System. J Neurosci. 2016;36: 11427–11434. doi: 10.1523/JNEUROSCI.2492-16.2016 - DOI - PMC - PubMed
-
- Ma DK, Marchetto MC, Guo JU, Ming GL, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci. 2010;13: 1338–1344. doi: 10.1038/nn.2672 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

