Unraveling historical introgression and resolving phylogenetic discord within Catostomus (Osteichthys: Catostomidae)
- PMID: 29879898
- PMCID: PMC5992631
- DOI: 10.1186/s12862-018-1197-y
Unraveling historical introgression and resolving phylogenetic discord within Catostomus (Osteichthys: Catostomidae)
Abstract
Background: Porous species boundaries can be a source of conflicting hypotheses, particularly when coupled with variable data and/or methodological approaches. Their impacts can often be magnified when non-model organisms with complex histories of reticulation are investigated. One such example is the genus Catostomus (Osteichthys, Catostomidae), a freshwater fish clade with conflicting morphological and mitochondrial phylogenies. The former is hypothesized as reflecting the presence of admixed genotypes within morphologically distinct lineages, whereas the latter is interpreted as the presence of distinct morphologies that emerged multiple times through convergent evolution. We tested these hypotheses using multiple methods, to including multispecies coalescent and concatenated approaches. Patterson's D-statistic was applied to resolve potential discord, examine introgression, and test the putative hybrid origin of two species. We also applied naïve binning to explore potential effects of concatenation.
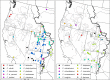
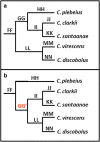
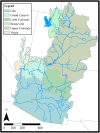
Results: We employed 14,007 loci generated from ddRAD sequencing of 184 individuals to derive the first highly supported nuclear phylogeny for Catostomus. Our phylogenomic analyses largely agreed with a morphological interpretation,with the exception of the placement of Xyrauchen texanus, which differs from both morphological and mitochondrial phylogenies. Additionally, our evaluation of the putative hybrid species C. columbianus revealed a lack introgression and instead matched the mitochondrial phylogeny. Furthermore, D-statistic tests clarified all discrepancies based solely on mitochondrial data, with agreement among topologies derived from concatenation and multispecies coalescent approaches. Extensive historic introgression was detected across six species-pairs. Potential endemism in the Virgin and Little Colorado Rivers was also apparent, and the former genus Pantosteus was derived as monophyletic, save for C. columbianus.
Conclusions: Complex reticulated histories detected herein support the hypothesis that introgression was responsible for conflicts that occurred within the mitochondrial phylogeny, and explains discrepancies found between it and previous morphological phylogenies. Additionally, the hybrid origin of C. columbianus was refuted, but with the caveat that more fine-grain sampling is still needed. Our diverse phylogenomic approaches provided largely concordant results, with naïve binning useful in exploring the single conflict. Considerable diversity was found within Catostomus across southwestern North America, with two drainages [Virgin River (UT) and Little Colorado River (AZ)] reflecting unique composition.
Keywords: Catostomus; Hybridization; Introgression; Patterson’s D-statistic; Phylogenetic incongruence; ddRAD.
Conflict of interest statement
Ethics approval and consent to participate
Arizona Game & Fish Department, Colorado Division of Wildlife, New Mexico Game & Fish Department, Utah Department of Wildlife Resources, Wyoming Game and Fish Department, U.S. National Park Service and U.S. Bureau of Reclamation contributed field expertise, specimens, technical assistance, collecting permits, funding and/or comments. Sampling procedures were approved under Institutional Animal Care and Use Committee permit 98-456R (Arizona State University) and 01-036A-01 (Colorado State University).
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Folk RA, Mandel JR, Freudenstein JV. Ancestral gene flow and parallel organellar genome capture result in extreme phylogenetic discord in a lineage of angiosperms. Syst Biol. 2017;66:320–337. - PubMed
-
- Lemmon EM, Lemmon AR. High-throughput genomic data in systematics and phylogenetics. Annu Rev Ecol Evol Syst. 2013;44:99–121. doi: 10.1146/annurev-ecolsys-110512-135822. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

