Hundreds of novel composite genes and chimeric genes with bacterial origins contributed to haloarchaeal evolution
- PMID: 29880023
- PMCID: PMC5992828
- DOI: 10.1186/s13059-018-1454-9
Hundreds of novel composite genes and chimeric genes with bacterial origins contributed to haloarchaeal evolution
Abstract
Background: Haloarchaea, a major group of archaea, are able to metabolize sugars and to live in oxygenated salty environments. Their physiology and lifestyle strongly contrast with that of their archaeal ancestors. Amino acid optimizations, which lowered the isoelectric point of haloarchaeal proteins, and abundant lateral gene transfers from bacteria have been invoked to explain this deep evolutionary transition. We use network analyses to show that the evolution of novel genes exclusive to Haloarchaea also contributed to the evolution of this group.
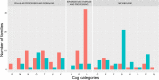
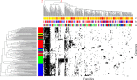
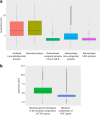
Results: We report the creation of 320 novel composite genes, both early in the evolution of Haloarchaea during haloarchaeal genesis and later in diverged haloarchaeal groups. One hundred and twenty-six of these novel composite genes derived from genetic material from bacterial genomes. These latter genes, largely involved in metabolic functions but also in oxygenic lifestyle, constitute a different gene pool from the laterally acquired bacterial genes formerly identified. These novel composite genes were likely advantageous for their hosts, since they show significant residence times in haloarchaeal genomes-consistent with a long phylogenetic history involving vertical descent and lateral gene transfer-and encode proteins with optimized isoelectric points.
Conclusions: Overall, our work encourages a systematic search for composite genes across all archaeal major groups, in order to better understand the origins of novel prokaryotic genes, and in order to test to what extent archaea might have adjusted their lifestyles by incorporating and recycling laterally acquired bacterial genetic fragments into new archaeal genes.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Origins of major archaeal clades correspond to gene acquisitions from bacteria.Nature. 2015 Jan 1;517(7532):77-80. doi: 10.1038/nature13805. Epub 2014 Oct 15. Nature. 2015. PMID: 25317564 Free PMC article.
-
Metabolic bacterial genes and the construction of high-level composite lineages of life.Trends Ecol Evol. 2015 Mar;30(3):127-9. doi: 10.1016/j.tree.2015.01.001. Epub 2015 Jan 16. Trends Ecol Evol. 2015. PMID: 25601290 Free PMC article.
-
Genomes in flux: the evolution of archaeal and proteobacterial gene content.Genome Res. 2002 Jan;12(1):17-25. doi: 10.1101/gr.176501. Genome Res. 2002. PMID: 11779827
-
Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world.Nucleic Acids Res. 2008 Dec;36(21):6688-719. doi: 10.1093/nar/gkn668. Epub 2008 Oct 23. Nucleic Acids Res. 2008. PMID: 18948295 Free PMC article. Review.
-
Protein based molecular markers provide reliable means to understand prokaryotic phylogeny and support Darwinian mode of evolution.Front Cell Infect Microbiol. 2012 Jul 26;2:98. doi: 10.3389/fcimb.2012.00098. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919687 Free PMC article. Review.
Cited by
-
The last universal common ancestor between ancient Earth chemistry and the onset of genetics.PLoS Genet. 2018 Aug 16;14(8):e1007518. doi: 10.1371/journal.pgen.1007518. eCollection 2018 Aug. PLoS Genet. 2018. PMID: 30114187 Free PMC article. Review.
-
Recovery of Lutacidiplasmatales archaeal order genomes suggests convergent evolution in Thermoplasmatota.Nat Commun. 2022 Jul 15;13(1):4110. doi: 10.1038/s41467-022-31847-7. Nat Commun. 2022. PMID: 35840579 Free PMC article.
-
Dynamic evolution of the mTHF gene family associated with primary metabolism across life.BMC Genomics. 2024 May 1;25(1):432. doi: 10.1186/s12864-024-10159-8. BMC Genomics. 2024. PMID: 38693486 Free PMC article.
-
Hikarchaeia demonstrate an intermediate stage in the methanogen-to-halophile transition.Nat Commun. 2020 Oct 30;11(1):5490. doi: 10.1038/s41467-020-19200-2. Nat Commun. 2020. PMID: 33127909 Free PMC article.
-
Ancestral Reconstructions Decipher Major Adaptations of Ammonia-Oxidizing Archaea upon Radiation into Moderate Terrestrial and Marine Environments.mBio. 2020 Oct 13;11(5):e02371-20. doi: 10.1128/mBio.02371-20. mBio. 2020. PMID: 33051370 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

