Proteomic analysis of Medulloblastoma reveals functional biology with translational potential
- PMID: 29880060
- PMCID: PMC5992829
- DOI: 10.1186/s40478-018-0548-7
Proteomic analysis of Medulloblastoma reveals functional biology with translational potential
Abstract
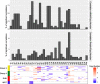
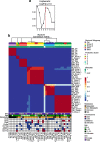
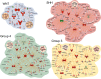
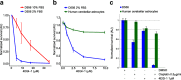
Genomic characterization has begun to redefine diagnostic classifications of cancers. However, it remains a challenge to infer disease phenotypes from genomic alterations alone. To help realize the promise of genomics, we have performed a quantitative proteomics investigation using Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) and 41 tissue samples spanning the 4 genomically based subgroups of medulloblastoma and control cerebellum. We have identified and quantitated thousands of proteins across these groups and find that we are able to recapitulate the genomic subgroups based upon subgroup restricted and differentially abundant proteins while also identifying subgroup specific protein isoforms. Integrating our proteomic measurements with genomic data, we calculate a poor correlation between mRNA and protein abundance. Using EPIC 850 k methylation array data on the same tissues, we also investigate the influence of copy number alterations and DNA methylation on the proteome in an attempt to characterize the impact of these genetic features on the proteome. Reciprocally, we are able to use the proteome to identify which genomic alterations result in altered protein abundance and thus are most likely to impact biology. Finally, we are able to assemble protein-based pathways yielding potential avenues for clinical intervention. From these, we validate the EIF4F cap-dependent translation pathway as a novel druggable pathway in medulloblastoma. Thus, quantitative proteomics complements genomic platforms to yield a more complete understanding of functional tumor biology and identify novel therapeutic targets for medulloblastoma.
Keywords: Medulloblastoma; Pediatric brain tumor; Proteomics; SILAC.
Conflict of interest statement
Ethics approval and consent to participate
Ethics approval has been obtained for the use of de-identified human tissue in this study.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



Similar articles
-
A Proteogenomic Approach to Understanding MYC Function in Metastatic Medulloblastoma Tumors.Int J Mol Sci. 2016 Oct 19;17(10):1744. doi: 10.3390/ijms17101744. Int J Mol Sci. 2016. PMID: 27775567 Free PMC article. Review.
-
Proteomic profiling of high risk medulloblastoma reveals functional biology.Oncotarget. 2015 Jun 10;6(16):14584-95. doi: 10.18632/oncotarget.3927. Oncotarget. 2015. PMID: 25970789 Free PMC article.
-
Proteomic profiling of medulloblastoma reveals novel proteins differentially expressed within each molecular subgroup.Clin Neurol Neurosurg. 2020 Sep;196:106028. doi: 10.1016/j.clineuro.2020.106028. Epub 2020 Jun 17. Clin Neurol Neurosurg. 2020. PMID: 32580068
-
Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma.J Clin Oncol. 2005 Dec 1;23(34):8853-62. doi: 10.1200/JCO.2005.02.8589. J Clin Oncol. 2005. PMID: 16314645
-
Medulloblastoma molecular dissection: the way toward targeted therapy.Curr Opin Oncol. 2013 Nov;25(6):674-81. doi: 10.1097/CCO.0000000000000008. Curr Opin Oncol. 2013. PMID: 24076581 Review.
Cited by
-
Workflow enabling deepscale immunopeptidome, proteome, ubiquitylome, phosphoproteome, and acetylome analyses of sample-limited tissues.Nat Commun. 2023 Apr 3;14(1):1851. doi: 10.1038/s41467-023-37547-0. Nat Commun. 2023. PMID: 37012232 Free PMC article.
-
Primary cilia contribute to the aggressiveness of atypical teratoid/rhabdoid tumors.Cell Death Dis. 2022 Sep 20;13(9):806. doi: 10.1038/s41419-022-05243-4. Cell Death Dis. 2022. PMID: 36127323 Free PMC article.
-
MeinteR: A framework to prioritize DNA methylation aberrations based on conformational and cis-regulatory element enrichment.Sci Rep. 2019 Dec 16;9(1):19148. doi: 10.1038/s41598-019-55453-8. Sci Rep. 2019. PMID: 31844073 Free PMC article.
-
Effective Inhibition of MYC-Amplified Group 3 Medulloblastoma Through Targeting EIF4A1.Cancer Manag Res. 2020 Dec 3;12:12473-12485. doi: 10.2147/CMAR.S278844. eCollection 2020. Cancer Manag Res. 2020. PMID: 33299354 Free PMC article.
-
Weighted Gene Co-Expression Network Analysis and Support Vector Machine Learning in the Proteomic Profiling of Cerebrospinal Fluid from Extraventricular Drainage in Child Medulloblastoma.Metabolites. 2022 Aug 5;12(8):724. doi: 10.3390/metabo12080724. Metabolites. 2022. PMID: 36005596 Free PMC article.
References
-
- Abe N, Watanabe T, Masaki T, Mori T, Sugiyama M, Uchimura H, Fujioka Y, Chiappetta G, Fusco A, Atomi Y. Pancreatic duct cell carcinomas express high levels of high mobility group I(Y) proteins. Cancer Res. 2000;60:3117–3122. - PubMed
-
- Abe N, Watanabe T, Sugiyama M, Uchimura H, Chiappetta G, Fusco A, Atomi Y. Determination of high mobility group I(Y) expression level in colorectal neoplasias: a potential diagnostic marker. Cancer Res. 1999;59:1169–1174. - PubMed
-
- Bhatia S, Baig NA, Timofeeva O, Pasquale EB, Hirsch K, MacDonald TJ, Dritschilo A, Lee YC, Henkemeyer M, Rood B. Knockdown of EphB1 receptor decreases medulloblastoma cell growth and migration and increases cellular radiosensitization. Oncotarget. 2015;6:8929–8946. doi: 10.18632/oncotarget.3369. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

