HuR regulates telomerase activity through TERC methylation
- PMID: 29880812
- PMCID: PMC5992219
- DOI: 10.1038/s41467-018-04617-7
HuR regulates telomerase activity through TERC methylation
Erratum in
-
Author Correction: HuR regulates telomerase activity through TERC methylation.Nat Commun. 2018 Jul 10;9(1):2721. doi: 10.1038/s41467-018-05213-5. Nat Commun. 2018. PMID: 29988036 Free PMC article.
Abstract
Telomerase consists of the catalytic protein TERT and the RNA TERC. Mutations in TERC are linked to human diseases, but the underlying mechanisms are poorly understood. Here we report that the RNA-binding protein HuR associates with TERC and promotes the assembly of the TERC/TERT complex by facilitating TERC C106 methylation. Dyskeratosis congenita (DC)-related TERC U100A mutation impair the association of HuR with TERC, thereby reducing C106 methylation. Two other TERC mutations linked to aplastic anemia and autosomal dominant DC, G107U, and GC107/108AG, likewise disrupt methylation at C106. Loss-of-HuR binding and hence lower TERC methylation leads to decreased telomerase activity and telomere shortening. Furthermore, HuR deficiency or mutation of mTERC HuR binding or methylation sites impair the renewal of mouse hematopoietic stem cells, recapitulating the bone marrow failure seen in DC. Collectively, our findings reveal a novel function of HuR, linking HuR to telomerase function and TERC-associated DC.
Conflict of interest statement
The authors declare no competing interests.
Figures


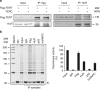

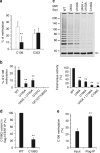
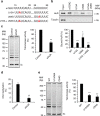
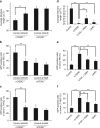
Similar articles
-
Heterozygous telomerase RNA mutations found in dyskeratosis congenita and aplastic anemia reduce telomerase activity via haploinsufficiency.Blood. 2004 Dec 15;104(13):3936-42. doi: 10.1182/blood-2004-05-1829. Epub 2004 Aug 19. Blood. 2004. PMID: 15319288
-
TERT and TERC mutations detected in cryptic dyskeratosis congenita suppress telomerase activity.Int J Lab Hematol. 2020 Jun;42(3):316-321. doi: 10.1111/ijlh.13176. Epub 2020 Mar 9. Int J Lab Hematol. 2020. PMID: 32150348
-
HuB and HuD repress telomerase activity by dissociating HuR from TERC.Nucleic Acids Res. 2021 Mar 18;49(5):2848-2858. doi: 10.1093/nar/gkab062. Nucleic Acids Res. 2021. PMID: 33589924 Free PMC article.
-
Dyskeratosis congenita: the diverse clinical presentation of mutations in the telomerase complex.Biochimie. 2008 Jan;90(1):122-30. doi: 10.1016/j.biochi.2007.07.017. Epub 2007 Jul 31. Biochimie. 2008. PMID: 17825470 Review.
-
Dyskeratosis congenita: telomerase, telomeres and anticipation.Curr Opin Genet Dev. 2005 Jun;15(3):249-57. doi: 10.1016/j.gde.2005.04.004. Curr Opin Genet Dev. 2005. PMID: 15917199 Review.
Cited by
-
How RNAi machinery enters the world of telomerase.Cell Cycle. 2019 May;18(10):1056-1067. doi: 10.1080/15384101.2019.1609834. Epub 2019 May 7. Cell Cycle. 2019. PMID: 31014212 Free PMC article. Review.
-
Interaction between HuR and circPABPN1 Modulates Autophagy in the Intestinal Epithelium by Altering ATG16L1 Translation.Mol Cell Biol. 2020 Feb 27;40(6):e00492-19. doi: 10.1128/MCB.00492-19. Print 2020 Feb 27. Mol Cell Biol. 2020. PMID: 31932481 Free PMC article.
-
Molecular Markers of Telomerase Complex for Patients with Pituitary Adenoma.Brain Sci. 2022 Jul 25;12(8):980. doi: 10.3390/brainsci12080980. Brain Sci. 2022. PMID: 35892421 Free PMC article.
-
Transcription stress at telomeres leads to cytosolic DNA release and paracrine senescence.Nat Commun. 2024 May 14;15(1):4061. doi: 10.1038/s41467-024-48443-6. Nat Commun. 2024. PMID: 38744897 Free PMC article.
-
Telomerase in cancer- ongoing quest and future discoveries.Mol Biol Rep. 2025 Jan 25;52(1):161. doi: 10.1007/s11033-025-10251-6. Mol Biol Rep. 2025. PMID: 39862360 Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

