40 Years of Research Put p53 in Translation
- PMID: 29883412
- PMCID: PMC5977125
- DOI: 10.3390/cancers10050152
40 Years of Research Put p53 in Translation
Abstract
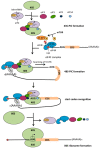
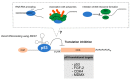
Since its discovery in 1979, p53 has shown multiple facets. Initially the tumor suppressor p53 protein was considered as a stress sensor able to maintain the genome integrity by regulating transcription of genes involved in cell cycle arrest, apoptosis and DNA repair. However, it rapidly came into light that p53 regulates gene expression to control a wider range of biological processes allowing rapid cell adaptation to environmental context. Among them, those related to cancer have been extensively documented. In addition to its role as transcription factor, scattered studies reported that p53 regulates miRNA processing, modulates protein activity by direct interaction or exhibits RNA-binding activity, thus suggesting a role of p53 in regulating several layers of gene expression not restricted to transcription. After 40 years of research, it appears more and more clearly that p53 is strongly implicated in translational regulation as well as in the control of the production and activity of the translational machinery. Translation control of specific mRNAs could provide yet unsuspected capabilities to this well-known guardian of the genome.
Keywords: cancer; p53; ribosome; translational control.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The multiple mechanisms that regulate p53 activity and cell fate.Nat Rev Mol Cell Biol. 2019 Apr;20(4):199-210. doi: 10.1038/s41580-019-0110-x. Nat Rev Mol Cell Biol. 2019. PMID: 30824861 Review.
-
Preferential translation of p53 target genes.RNA Biol. 2022;19(1):437-452. doi: 10.1080/15476286.2022.2048562. Epub 2021 Dec 31. RNA Biol. 2022. PMID: 35388737 Free PMC article.
-
p53 and translational control.Biochim Biophys Acta. 1996 Mar 18;1242(3):181-4. doi: 10.1016/0304-419x(95)00010-d. Biochim Biophys Acta. 1996. PMID: 8603071 Review.
-
Reversible induction of translational isoforms of p53 in glucose deprivation.Cell Death Differ. 2015 Jul;22(7):1203-18. doi: 10.1038/cdd.2014.220. Epub 2015 Feb 27. Cell Death Differ. 2015. PMID: 25721046 Free PMC article.
-
The tumor suppressors p53, p63, and p73 are regulators of microRNA processing complex.PLoS One. 2010 May 12;5(5):e10615. doi: 10.1371/journal.pone.0010615. PLoS One. 2010. PMID: 20485546 Free PMC article.
Cited by
-
The key target and role of betel nut in oral squamous cell carcinoma.BMC Oral Health. 2025 Jul 19;25(1):1212. doi: 10.1186/s12903-025-06558-2. BMC Oral Health. 2025. PMID: 40684141 Free PMC article.
-
Three-dimensional growth reveals fine-tuning of 5-lipoxygenase by proliferative pathways in cancer.Life Sci Alliance. 2023 Feb 27;6(5):e202201804. doi: 10.26508/lsa.202201804. Print 2023 May. Life Sci Alliance. 2023. PMID: 36849252 Free PMC article.
-
Nucleotide variations of TP53 exon 4 found in intracranial meningioma and in silico prediction of their significance.Mol Clin Oncol. 2019 Dec;11(6):563-572. doi: 10.3892/mco.2019.1936. Epub 2019 Oct 15. Mol Clin Oncol. 2019. PMID: 31692929 Free PMC article.
-
Shaping the regulation of the p53 mRNA tumour suppressor: the co-evolution of genetic signatures.BMC Cancer. 2019 Sep 13;19(1):915. doi: 10.1186/s12885-019-6118-y. BMC Cancer. 2019. PMID: 31519161 Free PMC article. Review.
-
The Ribosome Biogenesis-Cancer Connection.Cells. 2019 Jan 15;8(1):55. doi: 10.3390/cells8010055. Cells. 2019. PMID: 30650663 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

