The role of protein methyltransferases as potential novel therapeutic targets in squamous cell carcinoma of the head and neck
- PMID: 29884408
- PMCID: PMC6681457
- DOI: 10.1016/j.oraloncology.2018.04.014
The role of protein methyltransferases as potential novel therapeutic targets in squamous cell carcinoma of the head and neck
Erratum in
-
Corrigendum to "The role of protein methyltransferases as potential novel therapeutic targets in squamous cell carcinoma of the head and neck" [Oral Oncol. 81 (2018) 100-108].Oral Oncol. 2018 Nov;86:318. doi: 10.1016/j.oraloncology.2018.08.022. Epub 2018 Sep 7. Oral Oncol. 2018. PMID: 30201513 Free PMC article. No abstract available.
Abstract
Squamous cell carcinoma of the head and neck is a lethal disease with suboptimal survival outcomes and standard therapies with significant comorbidities. Whole exome sequencing data recently revealed an abundance of genetic and expression alterations in a family of enzymes known as protein methyltransferases in a variety of cancer types, including squamous cell carcinoma of the head and neck. These enzymes are mostly known for their chromatin-modifying functions through methylation of various histone substrates, though evidence supports their function also through methylation of non-histone substrates. This review summarizes the current knowledge on the function of protein methyltransferases in squamous cell carcinoma of the head and neck and highlights their promising potential as the next generation of therapeutic targets in this disease.
Keywords: EHMT2; EZH2; NSD1; NSD2; NSD3; PRMT1; PRMT5; Protein methylation; Protein methyltransferase; Squamous cell carcinoma of the head and neck.
Copyright © 2018 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Conflict of interest
The authors declare no relevant conflicts of interest.
Figures
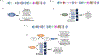

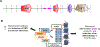

References
-
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2(5):401–4. 10.1158/2159-8290.CD-12-0095. - DOI - PMC - PubMed
-
- Somervaille T, Salamero O, Montesinos P, et al. 4060 Safety, Phamacokinetics (PK), Pharmacodynamics (PD) and preliminary activity in acute leukemia of ory-1001, a first-in-class inhibitor of lysine-specific histone demethylase 1A (LSD1/KDM1A): initial results from a first-in-human phase 1 study. Oral and poster abstracts, session: 616. Acute myeloid leukemia: novel therapy, excluding transplantation: poster III; December 5, 2016, San Diego.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

