Amniotic ectoderm expansion in mouse occurs via distinct modes and requires SMAD5-mediated signalling
- PMID: 29884675
- PMCID: PMC6053664
- DOI: 10.1242/dev.157222
Amniotic ectoderm expansion in mouse occurs via distinct modes and requires SMAD5-mediated signalling
Erratum in
-
Correction: Amniotic ectoderm expansion in mouse occurs via distinct modes and requires SMAD5-mediated signalling (doi: 10.1242/dev.157222).Development. 2018 Aug 2;145(15):dev169722. doi: 10.1242/dev.169722. Development. 2018. PMID: 30072497 Free PMC article. No abstract available.
Abstract
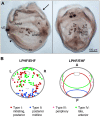
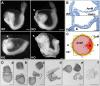
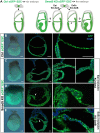
Upon gastrulation, the mammalian conceptus transforms rapidly from a simple bilayer into a multilayered embryo enveloped by its extra-embryonic membranes. Impaired development of the amnion, the innermost membrane, causes major malformations. To clarify the origin of the mouse amnion, we used single-cell labelling and clonal analysis. We identified four clone types with distinct clonal growth patterns in amniotic ectoderm. Two main types have progenitors in extreme proximal-anterior epiblast. Early descendants initiate and expand amniotic ectoderm posteriorly, while descendants of cells remaining anteriorly later expand amniotic ectoderm from its anterior side. Amniogenesis is abnormal in embryos deficient in the bone morphogenetic protein (BMP) signalling effector SMAD5, with delayed closure of the proamniotic canal, and aberrant amnion and folding morphogenesis. Transcriptomics of individual Smad5 mutant amnions isolated before visible malformations and tetraploid chimera analysis revealed two amnion defect sets. We attribute them to impairment of progenitors of the two main cell populations in amniotic ectoderm and to compromised cuboidal-to-squamous transition of anterior amniotic ectoderm. In both cases, SMAD5 is crucial for expanding amniotic ectoderm rapidly into a stretchable squamous sheet to accommodate exocoelom expansion, axial growth and folding morphogenesis.
Keywords: Amnion; Amnion fate map; BMP-SMAD; Chorion; Clonal analysis; Extra-embryonic ectoderm; Extra-embryonic–embryonic interface.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures





References
-
- Beddington R. S. P. and Lawson K. A. (1990). Clonal analysis of cell lineage. In Postimplantation Mammalian Embryos: A Practical Approach (ed. Copp A. J. and Cockroft D. L.), pp. 267-316. Oxford: IRL Press.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

