Substrate-Specific Differential Gene Expression and RNA Editing in the Brown Rot Fungus Fomitopsis pinicola
- PMID: 29884757
- PMCID: PMC6070754
- DOI: 10.1128/AEM.00991-18
Substrate-Specific Differential Gene Expression and RNA Editing in the Brown Rot Fungus Fomitopsis pinicola
Retraction in
-
Retraction for Wu et al., "Substrate-Specific Differential Gene Expression and RNA Editing in the Brown Rot Fungus Fomitopsis pinicola".Appl Environ Microbiol. 2021 Jul 27;87(16):e0033021. doi: 10.1128/AEM.00330-21. Epub 2021 Jul 27. Appl Environ Microbiol. 2021. PMID: 34313496 Free PMC article. No abstract available.
Retracted and republished in
-
Retracted and Republished from: "Substrate-Specific Differential Gene Expression and RNA Editing in the Brown Rot Fungus Fomitopsis pinicola".Appl Environ Microbiol. 2021 Jul 27;87(16):e0032921. doi: 10.1128/AEM.00329-21. Epub 2021 Jul 27. Appl Environ Microbiol. 2021. PMID: 34313495 Free PMC article.
Abstract
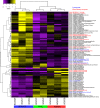
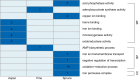
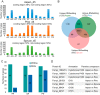
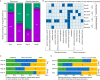
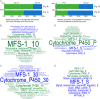
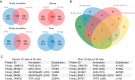
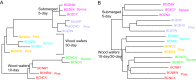
Wood-decaying fungi tend to have characteristic substrate ranges that partly define their ecological niche. Fomitopsis pinicola is a brown rot species of Polyporales that is reported on 82 species of softwoods and 42 species of hardwoods. We analyzed the gene expression levels and RNA editing profiles of F. pinicola from submerged cultures with ground wood powder (sampled at 5 days) or solid wood wafers (sampled at 10 and 30 days), using aspen, pine, and spruce substrates (aspen was used only in submerged cultures). Fomitopsis pinicola expressed similar sets of wood-degrading enzymes typical of brown rot fungi across all culture conditions and time points. Nevertheless, differential gene expression and RNA editing were observed across all pairwise comparisons of substrates and time points. Genes exhibiting differential expression and RNA editing encode diverse enzymes with known or potential function in brown rot decay, including laccase, benzoquinone reductase, aryl alcohol oxidase, cytochrome P450s, and various glycoside hydrolases. There was no overlap between differentially expressed and differentially edited genes, suggesting that these may provide F. pinicola with independent mechanisms for responding to different conditions. Comparing transcriptomes from submerged cultures and wood wafers, we found that culture conditions had a greater impact on global expression profiles than substrate wood species. In contrast, the suites of genes subject to RNA editing were much less affected by culture conditions. These findings highlight the need for standardization of culture conditions in studies of gene expression in wood-decaying fungi.IMPORTANCE All species of wood-decaying fungi occur on a characteristic range of substrates (host plants), which may be broad or narrow. Understanding the mechanisms that enable fungi to grow on particular substrates is important for both fungal ecology and applied uses of different feedstocks in industrial processes. We grew the wood-decaying polypore Fomitopsis pinicola on three different wood species, aspen, pine, and spruce, under various culture conditions. We examined both gene expression (transcription levels) and RNA editing (posttranscriptional modification of RNA, which can potentially yield different proteins from the same gene). We found that F. pinicola is able to modify both gene expression and RNA editing profiles across different substrate species and culture conditions. Many of the genes involved encode enzymes with known or predicted functions in wood decay. This work provides clues to how wood-decaying fungi may adjust their arsenal of decay enzymes to accommodate different host substrates.
Keywords: RNA editing; basidiomycetes; decay; lignocellulose; transcriptome.
Copyright © 2018 American Society for Microbiology.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

