The EGF/hnRNP Q1 axis is involved in tumorigenesis via the regulation of cell cycle-related genes
- PMID: 29884818
- PMCID: PMC5994831
- DOI: 10.1038/s12276-018-0101-6
The EGF/hnRNP Q1 axis is involved in tumorigenesis via the regulation of cell cycle-related genes
Abstract
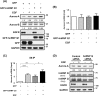
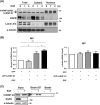
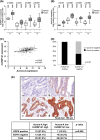
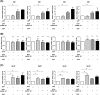
Heterogeneous nuclear ribonucleoprotein (hnRNP) Q1, an RNA-binding protein, has been implicated in many post-transcriptional processes, including RNA metabolism and mRNA splicing and translation. However, the role of hnRNP Q1 in tumorigenesis remains unclear. We previously performed RNA immunoprecipitation (RIP)-seq analysis to identify hnRNP Q1-interacting mRNAs and found that hnRNP Q1 targets a group of genes that are involved in mitotic regulation, including Aurora-A. Here, we demonstrate that altering the hnRNP Q1 level influences the expression of the Aurora-A protein, but not its mRNA. Stimulation with epidermal growth factor (EGF) enhances both binding between hnRNP Q1 and Aurora-A mRNA as well as the efficacy of the hnRNP Q1-induced translation of Aurora-A mRNA. The EGF/hnRNP Q1-induced translation of Aurora-A mRNA is mediated by the mTOR and ERK pathways. In addition, we show that hnRNP Q1 up-regulates the translation of a group of spindle assembly checkpoint (SAC) genes. hnRNP Q1 overexpression is positively correlated with the levels of Aurora-A and the SAC genes in human colorectal cancer tissues. In summary, our data suggest that hnRNP Q1 plays an important role in regulating the expression of a group of cell cycle-related genes. Therefore, it may contribute to tumorigenesis by up-regulating the translation of these genes in colorectal cancer.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

