Optimized protocol for combined PALM-dSTORM imaging
- PMID: 29884886
- PMCID: PMC5993819
- DOI: 10.1038/s41598-018-27059-z
Optimized protocol for combined PALM-dSTORM imaging
Abstract
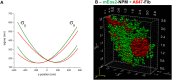
Multi-colour super-resolution localization microscopy is an efficient technique to study a variety of intracellular processes, including protein-protein interactions. This technique requires specific labels that display transition between fluorescent and non-fluorescent states under given conditions. For the most commonly used label types, photoactivatable fluorescent proteins and organic fluorophores, these conditions are different, making experiments that combine both labels difficult. Here, we demonstrate that changing the standard imaging buffer of thiols/oxygen scavenging system, used for organic fluorophores, to the commercial mounting medium Vectashield increased the number of photons emitted by the fluorescent protein mEos2 and enhanced the photoconversion rate between its green and red forms. In addition, the photophysical properties of organic fluorophores remained unaltered with respect to the standard imaging buffer. The use of Vectashield together with our optimized protocol for correction of sample drift and chromatic aberrations enabled us to perform two-colour 3D super-resolution imaging of the nucleolus and resolve its three compartments.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Hell SW. Far-field optical nanoscopy. 2010 23rd Annu. Meet. IEEE Photonics Soc. Photinics 2010. 2010;316:3–4. doi: 10.1109/PHOTONICS.2010.5698725. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

