Dissection of Influenza Infection In Vivo by Single-Cell RNA Sequencing
- PMID: 29886109
- PMCID: PMC7185763
- DOI: 10.1016/j.cels.2018.05.008
Dissection of Influenza Infection In Vivo by Single-Cell RNA Sequencing
Abstract
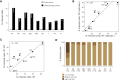
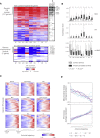
The influenza virus is a major cause of morbidity and mortality worldwide. Yet, both the impact of intracellular viral replication and the variation in host response across different cell types remain uncharacterized. Here we used single-cell RNA sequencing to investigate the heterogeneity in the response of lung tissue cells to in vivo influenza infection. Analysis of viral and host transcriptomes in the same single cell enabled us to resolve the cellular heterogeneity of bystander (exposed but uninfected) as compared with infected cells. We reveal that all major immune and non-immune cell types manifest substantial fractions of infected cells, albeit at low viral transcriptome loads relative to epithelial cells. We show that all cell types respond primarily with a robust generic transcriptional response, and we demonstrate novel markers specific for influenza-infected as opposed to bystander cells. These findings open new avenues for targeted therapy aimed exclusively at infected cells.
Keywords: bystander versus infected cells; immune and non-immune cell types; influenza infection in vivo; single-cell RNA sequencing.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

