i2b2 implemented over SMART-on-FHIR
- PMID: 29888095
- PMCID: PMC5961782
i2b2 implemented over SMART-on-FHIR
Abstract
Integrating Biology and the Bedside (i2b2) is the de-facto open-source medical tool for cohort discovery. Fast Healthcare Interoperability Resources (FHIR) is a new standard for exchanging health care information electronically. Substitutable Modular third-party Applications (SMART) defines the SMART-on-FHIR specification on how applications shall interface with Electronic Health Records (EHR) through FHIR. Related work made it possible to produce FHIR from an i2b2 instance or made i2b2 able to store FHIR datasets. In this paper, we extend i2b2 to search remotely into one or multiple SMART-on-FHIR Application Programming Interfaces (APIs). This enables the federation of queries, security, terminology mapping, and also bridges the gap between i2b2 and modern big-data technologies.
Figures

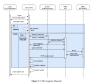


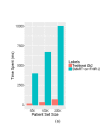

References
-
- Omri Gottesman, Helena Kuivaniemi, Gerard Tromp, W, Andrew Faucett, Rongling Li, Teri A. Manolio, Saskia C. Sanderson, Joseph Kannry, Randi Zinberg, Melissa A. Basford, et al. The electronic medical records and genomics (eMERGE) network: past, present, and future. Genetics in Medicine. 2013 Oct;15(10):761–771. - PMC - PubMed
-
- Georges De Moor, Mats Sundgren, Dipak Kalra, Andreas Schmidt, Martin Dugas, Brecht Claerhout, Toresin Karakoyun, Christian Ohmann, Pierre-Yves Lastic, Nadir Ammour, et al. Using electronic health records for clinical research: The case of the EHR4CR project. Journal of Biomedical Informatics. 2015 Feb;53:162–173. - PubMed
-
- Delaney Brendan C., Vasa Curcin, Anna Andreasson, Theodoros N. Arvanitis,, Hilde Bastiaens, Derek Corrigan, Ethier Jean-Francois, Olga Kostopoulou, Wolfgang Kuchinke, Mark McGilchrist, et al. Translational medicine and patient safety in Europe: TRANSFoRm-architecture for the learning health system in europe. BioMed Research International. 2015;2015:1–8. - PMC - PubMed
-
- George Hripcsak, Jon D. Duke, Nigam H. Shah, Christian G. Reich, Vojtech Huser, Martijn J, Schuemie Marc A, Suchard Rae, Woong Park, Ian Chi Kei Wong, Peter R. Rijnbeek, et al. Observational health data sciences and informatics (OHDSI): opportunities for observational researchers. Studies in health technology and informatics. 2015;216:574. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
