Dominant-negative IKZF1 mutations cause a T, B, and myeloid cell combined immunodeficiency
- PMID: 29889099
- PMCID: PMC6026000
- DOI: 10.1172/JCI98164
Dominant-negative IKZF1 mutations cause a T, B, and myeloid cell combined immunodeficiency
Abstract
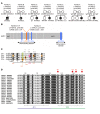
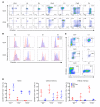
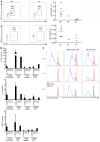
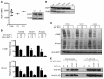
Ikaros/IKZF1 is an essential transcription factor expressed throughout hematopoiesis. IKZF1 is implicated in lymphocyte and myeloid differentiation and negative regulation of cell proliferation. In humans, somatic mutations in IKZF1 have been linked to the development of B cell acute lymphoblastic leukemia (ALL) in children and adults. Recently, heterozygous germline IKZF1 mutations have been identified in patients with a B cell immune deficiency mimicking common variable immunodeficiency. These mutations demonstrated incomplete penetrance and led to haploinsufficiency. Herein, we report 7 unrelated patients with a novel early-onset combined immunodeficiency associated with de novo germline IKZF1 heterozygous mutations affecting amino acid N159 located in the DNA-binding domain of IKZF1. Different bacterial and viral infections were diagnosed, but Pneumocystis jirovecii pneumonia was reported in all patients. One patient developed a T cell ALL. This immunodeficiency was characterized by innate and adaptive immune defects, including low numbers of B cells, neutrophils, eosinophils, and myeloid dendritic cells, as well as T cell and monocyte dysfunctions. Notably, most T cells exhibited a naive phenotype and were unable to evolve into effector memory cells. Functional studies indicated these mutations act as dominant negative. This defect expands the clinical spectrum of human IKZF1-associated diseases from somatic to germline, from haploinsufficient to dominant negative.
Keywords: Genetics; Immunology; Monocytes; Monogenic diseases; T cells.
Conflict of interest statement
Figures


References
-
- Molnár A, et al. The Ikaros gene encodes a family of lymphocyte-restricted zinc finger DNA binding proteins, highly conserved in human and mouse. J Immunol. 1996;156(2):585–592. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

