Analysis of protein-altering variants in telomerase genes and their association with MUC5B common variant status in patients with idiopathic pulmonary fibrosis: a candidate gene sequencing study
- PMID: 29891356
- PMCID: PMC6487850
- DOI: 10.1016/S2213-2600(18)30135-8
Analysis of protein-altering variants in telomerase genes and their association with MUC5B common variant status in patients with idiopathic pulmonary fibrosis: a candidate gene sequencing study
Abstract
Background: Idiopathic pulmonary fibrosis (IPF) risk has a strong genetic component. Studies have implicated variations at several loci, including TERT, surfactant genes, and a single nucleotide polymorphism at chr11p15 (rs35705950) in the intergenic region between TOLLIP and MUC5B. Patients with IPF who have risk alleles at rs35705950 have longer survival from the time of IPF diagnosis than do patients homozygous for the non-risk allele, whereas patients with shorter telomeres have shorter survival times. We aimed to assess whether rare protein-altering variants in genes regulating telomere length are enriched in patients with IPF homozygous for the non-risk alleles at rs35705950.
Methods: Between Nov 1, 2014, and Nov 1, 2016, we assessed blood samples from patients aged 40 years or older and of European ancestry with sporadic IPF from three international phase 3 clinical trials (INSPIRE, CAPACITY, ASCEND), one phase 2 study (RIFF), and US-based observational studies (Vanderbilt Clinical Interstitial Lung Disease Registry and the UCSF Interstitial Lung Disease Clinic registry cohorts) at the Broad Institute (Cambridge, MA, USA) and Human Longevity (San Diego, CA, USA). We also assessed blood samples from non-IPF controls in several clinical trials. We did whole-genome sequencing to assess telomere length and identify rare protein-altering variants, stratified by rs35705950 genotype. We also assessed rare functional variation in TERT exons and compared telomere length and disease progression across genotypes.
Findings: We assessed samples from 1510 patients with IPF and 1874 non-IPF controls. 30 (3%) of 1046 patients with an rs35705950 risk allele had a rare protein-altering variant in TERT compared with 34 (7%) of 464 non-risk allele carriers (odds ratio 0·40 [95% CI 0·24-0·66], p=0·00039). Subsequent analyses identified enrichment of rare protein-altering variants in PARN and RTEL1, and rare variation in TERC in patients with IPF compared with controls. We expanded our study population to provide a more accurate estimation of rare variant frequency in these four loci, and to calculate telomere length. The proportion of patients with at least one rare variant in TERT, PARN, TERC, or RTEL1 was higher in patients with IPF than in controls (149 [9%] of 1739 patients vs 205 [2%] of 8645 controls, p=2·44 × 10-8). Patients with IPF who had a variant in any of the four identified telomerase component genes had telomeres that were 3·69-16·10% shorter than patients without a variant in any of the four genes and had an earlier mean age of disease onset than patients without one or more variants (65·1 years [SD 7·8] vs 67·1 years [7·9], p=0·004). In the placebo arms of clinical trials, shorter telomeres were significantly associated with faster disease progression (1·7% predicted forced vital capacity per kb per year, p=0·002). Pirfenidone had treatment benefit regardless of telomere length (p=4·24 × 10-8 for telomere length lower than the median, p=0·0044 for telomere length greater than the median).
Interpretation: Rare protein-altering variants in TERT, PARN, TERC, and RTEL1 are enriched in patients with IPF compared with controls, and, in the case of TERT, particularly in individuals without a risk allele at the rs35705950 locus. This suggests that multiple genetic factors contribute to sporadic IPF, which might implicate distinct mechanisms of pathogenesis and disease progression.
Funding: Genentech, National Institutes of Health, Francis Family Foundation, Pulmonary Fibrosis Foundation, Nina Ireland Program for Lung Health, US Department of Veterans Affairs.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of Interest Statements
AD, ARA, JRA, BLY, CC, JaR, TRR, MN, TRB, MJB, JH, JeR, KM, KC, JT, AC, JV, WFF are employees of Genentech who hold stock and stock options in Roche. LHL is or has consulted for Genentech, Boehringer Ingelheim, Global Blood Therapeutics, and Bellerophon and has done disease state education for Genentech and Boehringer Ingelheim.
Figures


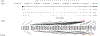

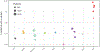



Comment in
-
Towards genetic reclassification of idiopathic pulmonary fibrosis.Lancet Respir Med. 2018 Aug;6(8):569-570. doi: 10.1016/S2213-2600(18)30173-5. Epub 2018 Jun 8. Lancet Respir Med. 2018. PMID: 29891357 No abstract available.
-
Advances in Interstitial Lung Disease Genetics.Am J Respir Crit Care Med. 2019 Jul 15;200(2):247-249. doi: 10.1164/rccm.201902-0471RR. Am J Respir Crit Care Med. 2019. PMID: 31014082 No abstract available.
References
-
- Richeldi L, Collard HR, Jones MG. Idiopathic pulmonary fibrosis. Lancet 2017; 389(10082): 1941–52. - PubMed
-
- Noble PW, Albera C, Bradford WZ, et al. Pirfenidone in patients with idiopathic pulmonary fibrosis (CAPACITY): two randomised trials. Lancet 2011; 377(9779): 1760–9. - PubMed
-
- King TE Jr., Bradford WZ, Castro-Bernardini S, et al. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med 2014; 370(22): 2083–92. - PubMed
-
- Richeldi L, du Bois RM, Raghu G, et al. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med 2014; 370(22): 2071–82. - PubMed
-
- Raghu G, Rochwerg B, Zhang Y, et al. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline: Treatment of Idiopathic Pulmonary Fibrosis. An Update of the 2011 Clinical Practice Guideline. Am J Respir Crit Care Med 2015; 192(2): e3–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

